





Acta Veterinaria et Zootechnica Sinica ›› 2025, Vol. 56 ›› Issue (9): 4432-4451.doi: 10.11843/j.issn.0366-6964.2025.09.025
• Animal Biotechnology and Reproduction • Previous Articles Next Articles
WANG Rui( ), HENG Nuo, HU Yingfan, WANG Huan, ZHU Ni, HE Wei, XUAN Xiuli, HU Zhihui, XIONG Keng, GONG Jianfei, HAO Haisheng, ZHU Huabin, ZHAO Shanjiang*(
), HENG Nuo, HU Yingfan, WANG Huan, ZHU Ni, HE Wei, XUAN Xiuli, HU Zhihui, XIONG Keng, GONG Jianfei, HAO Haisheng, ZHU Huabin, ZHAO Shanjiang*( )
)
Received:2025-03-11
Online:2025-09-23
Published:2025-09-30
Contact:
ZHAO Shanjiang
E-mail:ruiwang202010@163.com;zhaoshanjiang@caas.cn
CLC Number:
WANG Rui, HENG Nuo, HU Yingfan, WANG Huan, ZHU Ni, HE Wei, XUAN Xiuli, HU Zhihui, XIONG Keng, GONG Jianfei, HAO Haisheng, ZHU Huabin, ZHAO Shanjiang. Mechanisms of Developmental Competence Differences of Oocytes from Different Grades of Bovine Cumulus-Oocyte Complexes after in Vitro Maturation[J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(9): 4432-4451.
Table 1
Primers for the ddPCR and qRT-PCR"
| 引物名称 Primer name | 引物序列(5′→3′) Sequence | 产物长度/bp PCR size | 退火温度/℃ Annealing temperature |
| GAPDH-Forward | CACCCTCAAGATTGTCAGCA | 103 | 58 |
| GAPDH-Reverse | GGTCATAAGTCCCTCCACGA | ||
| PKM-Forward | CCCACAGTCGGGATAACCTTG | 127 | 58 |
| PKM-Reverse | TCCAGAGGAACTCCGACG | ||
| IDH3α-Forward | TGCAATCTTCGAGTCGGTTCA | 130 | 60 |
| IDH3α-Reverse | CTTCGCAGCGTGGTCAAAAA | ||
| β-actin-Forward | GCCCTGAGGCTCTCTTCCA | 101 | 60 |
| β-actin-Reverse | GCGGATGTCGACGTCACA | ||
| NDUFA4L2-Forward | CGATGATCGGCTTCATTGGC | 103 | 59 |
| NDUFA4L2-Reverse | GGGTTGTTCTTTCGGTCCCA |
Table 2
Comparison of in vitro developmental ability of oocytes from first-, second- and third-, fourth-grade COCs"
| 组别 Group | 体外培养总数/枚 Total number of embryos in vitro | 卵裂数/枚(%±SEM) Cleavage number | 囊胚数/枚(%±SEM) Number of blastocysts |
| 一二级COCs First- and second-grade COCs | 365 | 285(78.09±3.01)a | 93(32.56±4.33)a |
| 三四级COCs Third- and fourth-grade COCs | 317 | 192(60.59±6.54)b | 43(22.11±2.79)b |
Table 3
Smart-seq data quality analysis"
| 样本 Sample | Raw reads数量 Raw reads number | Clean reads数量 Clean reads number | Effective reads比率/% Effective reads rate | Mapped reads比率/% Mapped reads rate | Q30值/% Q30 value |
| GV1-1 | 45 674 378 | 43 982 102 | 96.29 | 94.42 | 95.75 |
| GV1-2 | 51 129 184 | 49 398 606 | 96.62 | 94.28 | 95.90 |
| GV1-3 | 54 554 672 | 52 536 610 | 96.30 | 93.81 | 95.99 |
| GV3-1 | 67 726 306 | 65 549 142 | 96.79 | 94.24 | 95.85 |
| GV3-2 | 48 507 034 | 46 525 878 | 95.92 | 93.56 | 95.79 |
| GV3-3 | 46 170 534 | 44 167 230 | 95.66 | 93.49 | 95.50 |
| MII1-1 | 54 309 472 | 53 026 116 | 97.64 | 95.64 | 96.20 |
| MII1-2 | 52 572 712 | 50 965 326 | 96.94 | 94.99 | 95.96 |
| MII1-3 | 58 974 948 | 57 600 648 | 97.67 | 95.27 | 96.35 |
| MII3-1 | 61 658 056 | 59 849 842 | 97.07 | 95.19 | 96.09 |
| MII3-2 | 65 222 820 | 62 237 638 | 95.42 | 93.64 | 95.80 |
| MII3-3 | 69 933 254 | 67 284 420 | 96.21 | 93.76 | 95.96 |

Fig. 1
PCA analysis of bovine oocyte transcripts A.PCA analysis of transcripts of first-, second-grade oocytes and third-, fourth-grade oocytes before maturation; B. PCA analysis of transcripts of first-, second-grade oocytes and third-, fourth-grade oocytes after maturation(22-24 h in vitro)"
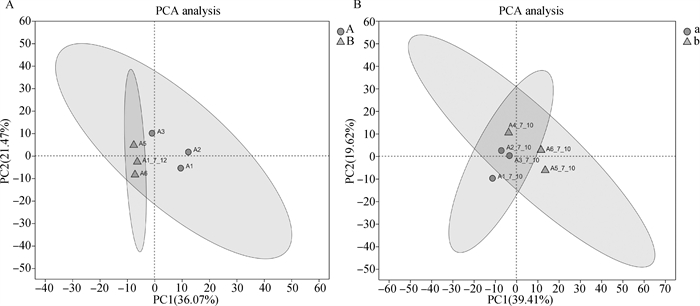

Fig. 2
Comparison of significant DEGs between groups before and after maturation(22-24 h in vitro) A.Volcano plot comparing significant DEGs between groups before maturation; B. Volcano plot comparing significant DEGs between groups after maturation(22-24 h in vitro); C. Veen plot comparing significant DEGs between groups before maturation (left image) and after maturation (right image)"
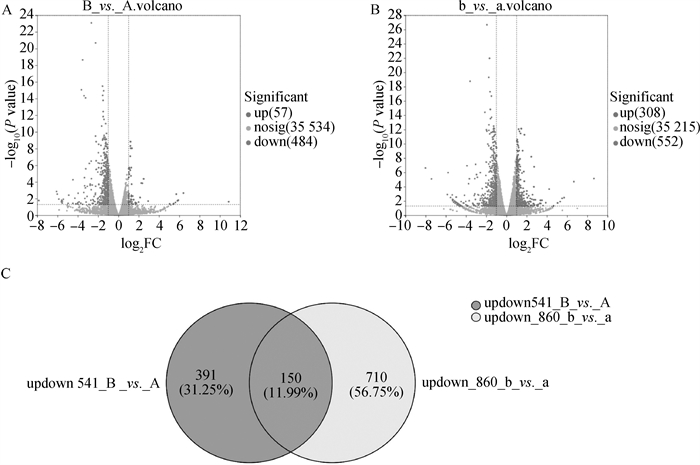

Fig. 3
Significant DEGs GO enrichment plot A. GO enrichment plot of significant DEGs in first-, second-grade oocytes and third-, fourth-grade oocytes before maturation; B. GO enrichment plot of significant DEGs in first-, second-grade oocytes and third-, fourth-grade oocytes after maturation (22-24 h in vitro)"
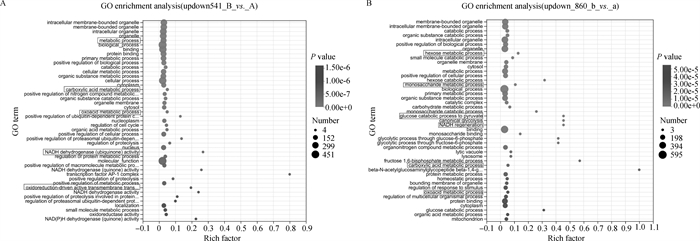

Fig. 4
Significant DEGs KEGG enrichment plot A.KEGG enrichment plot of significant DEGs in first-, second-grade oocytes and third-, fourth-grade oocytes before maturation; B. KEGG enrichment plot of significant DEGs in first-, second-grade oocytes and third-, fourth-grade oocytes after maturation(22-24 h in vitro)"
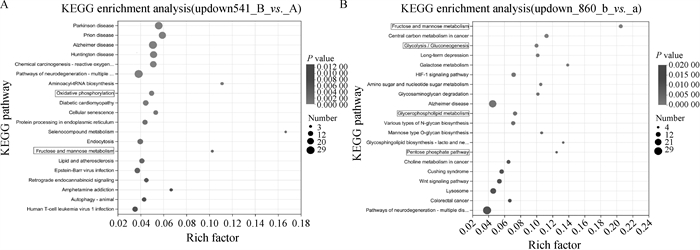
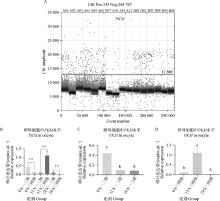
Fig. 5
PKM gene digital PCR verification results A. Copy number scatter plots of PKM gene in DNA solutions of first-, sceond- and third-, fourth-grade oocytes matured in vitro for 0, 12, and 22-24 h; B. Copy number bar chart of PKM gene in DNA solutions of first-, sceond- and third-, fourth-grade oocytes matured in vitro for 0, 12, and 22-24 h; C. Copy number bar chart of PKM gene in DNA solution of first-sceond-grade oocytes matured in vitro for 0, 12, and 22-24 h; D. Copy number bar chart of PKM gene in DNA solution of third- and fourth-grade oocytes matured in vitro for 0, 12, and 22-24 h"
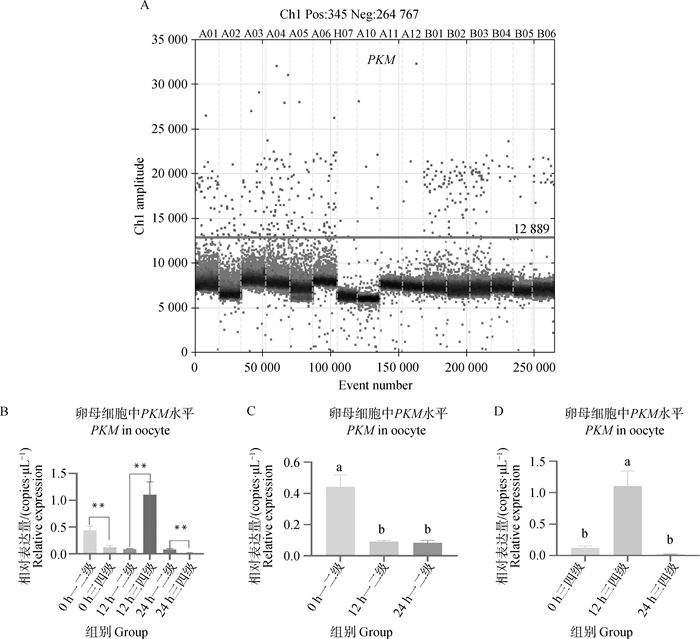
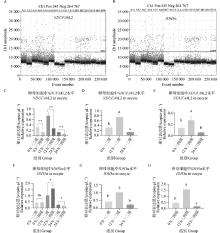
Fig. 6
NDUFA4L2 and IDH3α genes digital PCR verification results A. Copy number scatter plots of NDUFA4L2 gene in DNA solutions of first-, second- and third-, fourth-grade oocytes matured in vitro for 0, 12, and 22-24 h; B. Copy number scatter plots of IDH3α gene in DNA solutions of first-, second- and third-, fourth-grade oocytes matured in vitro for 0, 12, and 22-24 h; C. Copy number bar chart of NDUFA4L2 gene in DNA solutions of first-, second- and third-, fourth-grade oocytes matured in vitro for 0, 12, and 22-24 h; D. Copy number bar chart of NDUFA4L2 gene in DNA solutions of first-, second-grade oocytes matured in vitro for 0, 12, and 22-24 h; E. Copy number bar chart of NDUFA4L2 gene in DNA solutions of third- and fourth-grade oocytes matured in vitro for 0, 12, and 22-24 h; F. Copy number bar chart of IDH3α gene in DNA solutions of first-, second- and third-, fourth-grade oocytes matured in vitro for 0, 12, and 22-24 h; G. Copy number bar chart of IDH3α gene in DNA solutions of first-, second-grade oocytes matured in vitro for 0, 12, and 22-24 h; H. Copy number bar chart of IDH3α gene in DNA solutions of third- and fourth-grade oocytes matured in vitro for 0, 12, and 22-24 h"
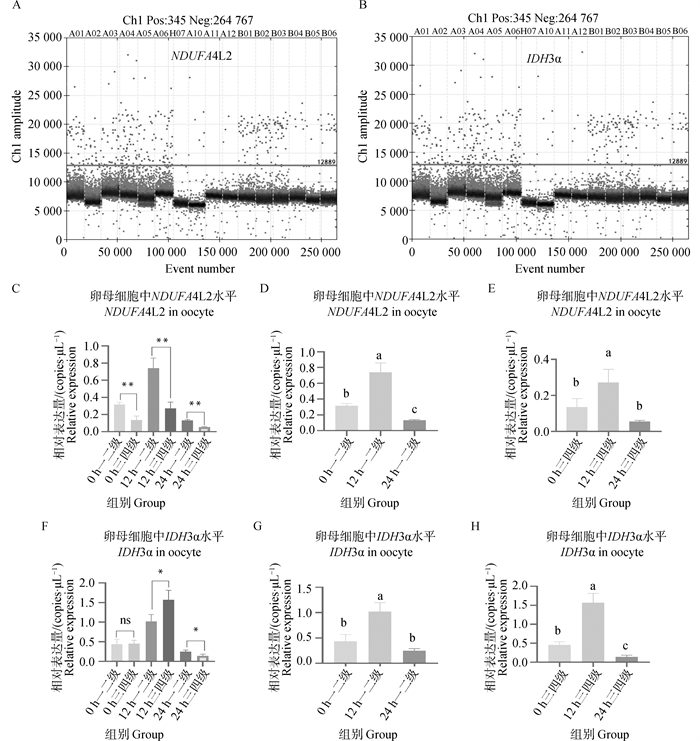
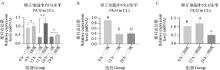
Fig. 7
PKM gene qRT-PCR verification results A. qRT-PCR detection results of PKM in first-, sceond- and third-, fourth-grade cumulus cells matured in vitro at 0, 12, and 22-24 h(n=3);B. qRT-PCR detection results of PKM in first-, second-grade cumulus cells matured in vitro at 0, 12, and 22-24 h(n=3);C. qRT-PCR detection results of PKM in third- and fourth-grade cumulus cells matured in vitro at 0, 12, and 22-24 h(n=3)"
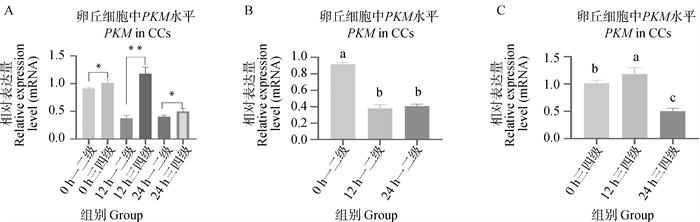

Fig. 8
IDH3α and NDUFA4L2 genes qRT-PCR verification results A. qRT-PCR detection results of IDH3α in first-, second- and third-, fourth-grade cumulus cells matured in vitro at 0, 12, and 22-24 h(n=3);B. qRT-PCR detection results of IDH3α in first-, second-grade cumulus cells matured in vitro at 0, 12, and 22-24 h(n=3);C. qRT-PCR detection results of IDH3α in third- and fourth-grade cumulus cells matured in vitro at 0, 12, and 22-24 h(n=3);D. qRT-PCR detection results of NDUFA4L2 in first-, second- and third-, fourth-grade cumulus cells matured in vitro at 0, 12, and 22-24 h(n=3);E. qRT-PCR detection results of NDUFA4L2 in first-, second-grade cumulus cells matured in vitro at 0, 12, and 22-24 h(n=3);F. qRT-PCR detection results of NDUFA4L2 in third- and fourth-grade cumulus cells matured in vitro at 0, 12, and 22-24 h(n=3)"
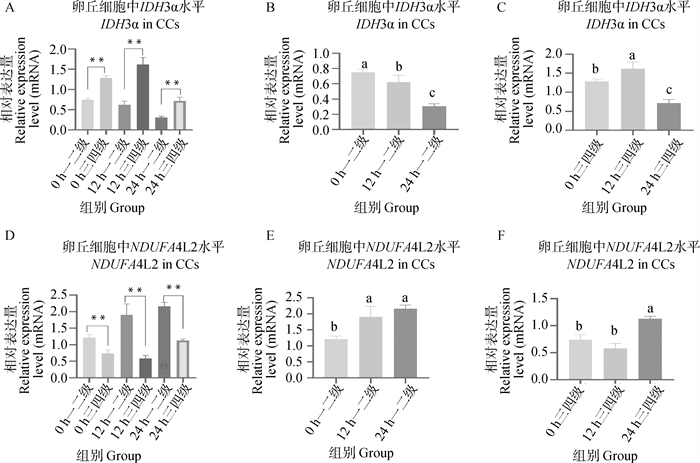

Fig. 9
OPLS-DA and displacement assay analysis of metabolites in mature culture medium A. OPLS-DA analysis of mature culture medium for first- and second-stage (TR1) and third- and fourth-stage (TR3) cumulus oocyte complexes in positive ion mode; B. Displacement assay analysis of mature culture medium for first- and second-stage (TR1) and third- and fourth-stage (TR3) cumulus oocyte complexes in positive ion mode; C. OPLS-DA analysis of mature culture medium for first- and second-stage (TR1) and third- and fourth-stage (TR3) cumulus oocyte complexes in negative ion mode; D. Displacement assay analysis of mature culture medium for first- and second-stage (TR1) and third- and fourth-stage (TR3) cumulus oocyte complexes in negative ion mode"

Fig. 10
The significant differential metabolites between groups A. The volcano plot of significant differential metabolites in in vitro maturation culture medium of first- and second-grade (TR1) and third- and fourth-grade (TR3) cumulus oocyte complexes under positive and negative ion modes; B. The column chart of significant differential metabolites in in vitro maturation culture medium of first- and second-grade (TR1) and third- and fourth-grade (TR3) cumulus oocyte complexes under positive and negative ion modes"
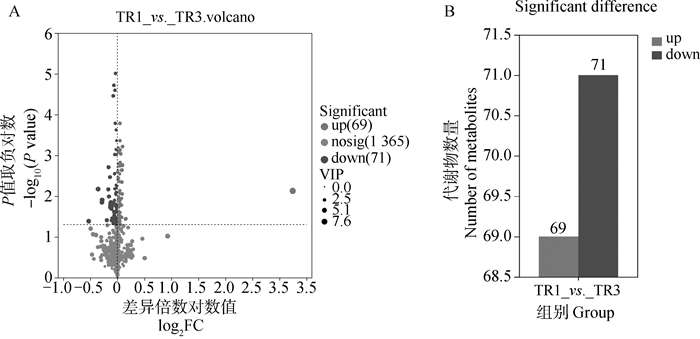

Fig. 11
Enrichment analysis of significant differential metabolites A. Pathway analysis of significantly upregulated differential metabolites in the maturation buffer of first-, second- and third-, fourth-grade cumulus oocyte complexes; B. Pathway analysis of significantly downregulated differential metabolites in the maturation buffer of first-, second- and third-, fourth-grade cumulus oocyte complexes"
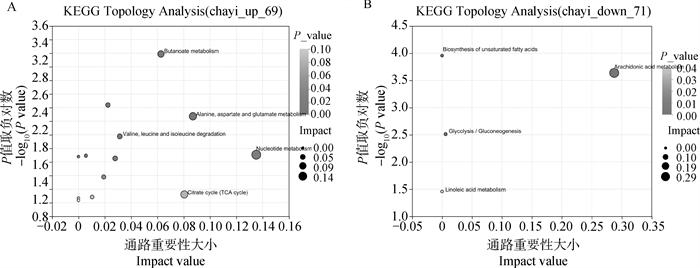

Fig. 13
Analysis of relative abundance of differential metabolites in oocyte maturation fluid A. Relative abundance map of α-D-glucose in oocyte maturation fluid; B. Relative abundance map of pyruvate in oocyte maturation fluid; C. Relative abundance map of lactic acid in oocyte maturation fluid"
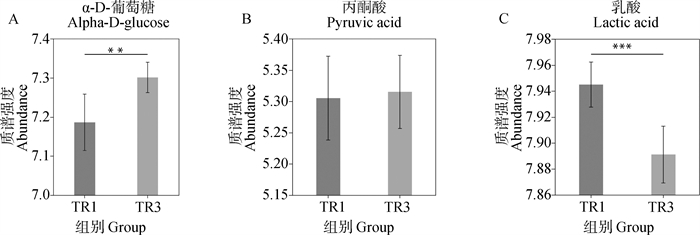
Table 4
The effect of adding sodium pyruvate on the developmental ability of third- and fourth-grade oocytes"
| 组别 Group | 培养COCs总数/枚 Total number of COCs cultivated | 卵裂数/枚 ((mean±SEM)%) Cleavage number | 囊胚数/枚 ((mean±SEM)%) Number of blastocysts |
| 一二级COCs First- and second-grade COCs | 412 | 313 (75.15±4.46) | 105 (33.02±2.76) |
| 三四级COCs+丙酮酸钠 Third- and fourth-grade COCs +sodium pyruvate | 322 | 234 (72.54±4.79)a | 71 (30.63±4.15)a |
| 三四级COCs Third- and fourth-grade COCs | 311 | 192 (61.71±4.75)b | 44 (22.72±2.39)b |
| 1 | VIANA J H . 2023 Statistics of embryo production and transfer in domestic farm animals[J]. Embr Tech Newsl, 2024, 42 (4): 31- 46. |
| 2 | KRISHER R L . The effect of oocyte quality on development[J]. J Anim Sci, 2004, 82 (E-Suppl): E14- E23. |
| 3 |
HOMER H A . The role of oocyte quality in explaining "Unexplained" infertility[J]. Semin Reprod Med, 2020, 38 (1): 21- 28.
doi: 10.1055/s-0040-1721377 |
| 4 |
GALLO A , ESPOSITO M C , BONI R , et al. Oocyte quality assessment in marine invertebrates: a novel approach by fluorescence spectroscopy[J]. Biol Res, 2022, 55 (1): 34.
doi: 10.1186/s40659-022-00403-4 |
| 5 |
JEENA L M , KUMAR D , RAHANGDALE S , et al. Effect of cumulus cells of cumulus-oocyte complexes on in vitro maturation, embryonic developmental and expression pattern of apoptotic genes after in vitro fertilization in water buffalo (Bubalus bubalis)[J]. Anim Biotechnol, 2020, 31 (2): 135- 141.
doi: 10.1080/10495398.2018.1554580 |
| 6 |
DE LOOS F , VAN VLIET C , VAN MAURIK P , et al. Morphology of immature bovine oocytes[J]. Gamete Res, 1989, 24 (2): 197- 204.
doi: 10.1002/mrd.1120240207 |
| 7 | RODRIGUEZ-VARELA C , HERRAIZ S , LABARTA E . Mitochondrial enrichment in infertile patients: a review of different mitochondrial replacement therapies[J]. Ther Adv Reprod Health, 2021, 15, 1343248568. |
| 8 |
HAO X , ZHAO J , RODRIGUEZ-WALLBERG K A . Comprehensive atlas of mitochondrial distribution and dynamics during oocyte maturation in mouse models[J]. Biomark Res, 2024, 12 (1): 125.
doi: 10.1186/s40364-024-00672-z |
| 9 |
YUN Y , LEE S , SO C , et al. Oocyte development and quality in young and old mice following exposure to atrazine[J]. Environ Health Perspect, 2022, 130 (11): 117007.
doi: 10.1289/EHP11343 |
| 10 |
PETERS A E , FORD E A , ROMAN S D , et al. Impact of Bisphenol A and its alternatives on oocyte health: a scoping review[J]. Hum Reprod Update, 2024, 30 (6): 653- 691.
doi: 10.1093/humupd/dmae025 |
| 11 | 安卓, 谢聪聪, 赵明, 等. 线粒体对卵母细胞发育的影响[J]. 生殖医学杂志, 2021, 30 (11): 1533- 1537. |
| AN Z , XIE C C , ZHAO M , et al. Eeffects of mitochondria on oocyte development[J]. Journal Reproductive Medicine, 2021, 30 (11): 1533- 1537. | |
| 12 |
XIONG Y Y , ZHU H Y , SHI R J , et al. Regulation of glucose metabolism: Effects on oocyte, preimplantation embryo, assisted reproductive technology and embryonic stem cell[J]. Heliyon, 2024, 10 (19): e38551.
doi: 10.1016/j.heliyon.2024.e38551 |
| 13 |
KOBAYASHI H , YOSHIMOTO C , MATSUBARA S , et al. Altered energy metabolism, mitochondrial dysfunction, and redox imbalance influencing reproductive performance in granulosa cells and oocyte during aging[J]. Reprod Sci, 2024, 31 (4): 906- 916.
doi: 10.1007/s43032-023-01394-7 |
| 14 |
NAHAR A , BECKER J , PASQUARIELLO R , et al. FGF2, LIF, and IGF-1 supplementation improves mouse oocyte in vitro maturation via increased glucose metabolismdagger[J]. Biol Reprod, 2024, 110 (4): 672- 683.
doi: 10.1093/biolre/ioae014 |
| 15 |
ZHANG Q , REN J , WANG F , et al. Mitochondrial and glucose metabolic dysfunctions in granulosa cells induce impaired oocytes of polycystic ovary syndrome through Sirtuin 3[J]. Free Radic Biol Med, 2022, 187, 1- 16.
doi: 10.1016/j.freeradbiomed.2022.05.010 |
| 16 |
PICELLI S , FARIDANI O R , BJORKLUND A K , et al. Full-length RNA-seq from single cells using Smart-seq2[J]. Nat Protoc, 2014, 9 (1): 171- 181.
doi: 10.1038/nprot.2014.006 |
| 17 |
OU Z , DU J , LIU N , et al. The impact of low oocyte maturity ratio on blastocyst euploidy rate: a matched retrospective cohort study[J]. Contracept Reprod Med, 2024, 9 (1): 41.
doi: 10.1186/s40834-024-00303-w |
| 18 |
PARRELLA A , IRANI M , KEATING D , et al. High proportion of immature oocytes in a cohort reduces fertilization, embryo development, pregnancy and live birth rates following ICSI[J]. Reprod Biomed Online, 2019, 39 (4): 580- 587.
doi: 10.1016/j.rbmo.2019.06.005 |
| 19 |
XIE H L , WANG Y B , JIAO G Z , et al. Effects of glucose metabolism during in vitro maturation on cytoplasmic maturation of mouse oocytes[J]. Sci Rep, 2016, 6, 20764.
doi: 10.1038/srep20764 |
| 20 |
ALVAREZ G M , BARRIOS E M , ELIA E , et al. Effects of gonadotrophins and insulin on glucose uptake in the porcine cumulus-oocyte complex during ⅣM[J]. Reprod Fertil Dev, 2019, 31 (8): 1353- 1359.
doi: 10.1071/RD18321 |
| 21 |
ABDULHASAN M K , LI Q , DAI J , et al. CoQ10 increases mitochondrial mass and polarization, ATP and Oct4 potency levels, and bovine oocyte MII during ⅣM while decreasing AMPK activity and oocyte death[J]. J Assist Reprod Genet, 2017, 34 (12): 1595- 1607.
doi: 10.1007/s10815-017-1027-y |
| 22 |
ZHANG Y , JIANG X , SONG X , et al. Mendelian randomization and multi-omics approach analyses reveal impaired glucose metabolism and oxidative phosphorylation in visceral adipose tissue of women with polycystic ovary syndrome[J]. Hum Reprod, 2024, 39 (12): 2785- 2797.
doi: 10.1093/humrep/deae244 |
| 23 |
BOLAND N I , HUMPHERSON P G , LEESE H J , et al. The effect of glucose metabolism on murine follicle development and steroidogenesis in vitro[J]. Hum Reprod, 1994, 9 (4): 617- 623.
doi: 10.1093/oxfordjournals.humrep.a138559 |
| 24 |
PROCHOWNIK E V , WANG H . The metabolic fates of pyruvate in normal and neoplastic cells[J]. Cells, 2021, 10 (4): 762.
doi: 10.3390/cells10040762 |
| 25 | LI Y R , PENG R R , GAO W Y , et al. The ubiquitin ligase KBTBD8 regulates PKM1 levels via Erk1/2 and Aurora A to ensure oocyte quality[J]. Aging (Albany NY), 2019, 11 (4): 1110- 1128. |
| 26 | LIU Z , CHAILLOU T , SANTOS A E , et al. Mitochondrial NDUFA4L2 is a novel regulator of skeletal muscle mass and force[J]. FASEB J, 2021, 35 (12): e22010. |
| 27 |
CHEN Y C , LI J Y , LI C J , et al. Luteal phase ovarian stimulation versus follicular phase ovarian stimulation results in different human cumulus cell genes expression: A pilot study[J]. Int J Med Sci, 2021, 18 (7): 1600- 1608.
doi: 10.7150/ijms.55955 |
| 28 |
HOU J Y , WANG X L , CHANG H J , et al. PTBP1 crotonylation promotes colorectal cancer progression through alternative splicing-mediated upregulation of the PKM2 gene[J]. J Transl Med, 2024, 22 (1): 995.
doi: 10.1186/s12967-024-05793-5 |
| 29 |
WEN J , WANG G L , YUAN H J , et al. Effects of glucose metabolism pathways on nuclear and cytoplasmic maturation of pig oocytes[J]. Sci Rep, 2020, 10 (1): 2782.
doi: 10.1038/s41598-020-59709-6 |
| 30 |
SUTTON-MCDOWALL M L , GILCHRIST R B , THOMPSON J G . The pivotal role of glucose metabolism in determining oocyte developmental competence[J]. Reproduction, 2010, 139 (4): 685- 695.
doi: 10.1530/REP-09-0345 |
| 31 | 赵昍朋, 李宝山, 程东凯. 颗粒细胞糖脂代谢对卵母细胞发育影响的研究进展[J]. 妇儿健康导刊, 2024, 3 (15): 22- 26. |
| ZHAO X P , LI B S , CHENG D K . Research progress on the effects of granulosa cell glucose and lipid metabolism on oocyte development[J]. Fuer Jiankang Daokan, 2024, 3 (15): 22- 26. | |
| 32 |
DALTON C M , SZABADKAI G , CARROLL J . Measurement of ATP in single oocytes: impact of maturation and cumulus cells on levels and consumption[J]. J Cell Physiol, 2014, 229 (3): 353- 361.
doi: 10.1002/jcp.24457 |
| 33 | SHI L , WANG H , ZHU S , et al. Multi-omics reveal the metabolic changes in cumulus cells during aging[J]. Cell Prolif, 2025, e70014. |
| 34 |
ZHANG Q , HAO J X , LIU B W , et al. Supplementation of mitochondria from endometrial mesenchymal stem cells improves oocyte quality in aged mice[J]. Cell Prolif, 2023, 56 (3): e13372.
doi: 10.1111/cpr.13372 |
| 35 |
MEI N H , GUO S M , ZHOU Q , et al. H3K4 methylation promotes expression of mitochondrial dynamics regulators to ensure oocyte quality in mice[J]. Adv Sci (Weinh), 2023, 10 (12): e2204794.
doi: 10.1002/advs.202204794 |
| 36 |
ZHANG Y , LIU L , YIN T L , et al. Follicular metabolic changes and effects on oocyte quality in polycystic ovary syndrome patients[J]. Oncotarget, 2017, 8 (46): 80472- 80480.
doi: 10.18632/oncotarget.19058 |
| 37 |
MOORE S G , FAIR T , LONERGAN P , et al. Genetic merit for fertility traits in Holstein cows: Ⅳ. Transition period, uterine health, and resumption of cyclicity[J]. J Dairy Sci, 2014, 97 (5): 2740- 2752.
doi: 10.3168/jds.2013-7278 |
| 38 |
CAI X , NG C P , JONES O , et al. Lactate activates the mitochondrial electron transport chain independently of its metabolism[J]. Mol Cell, 2023, 83 (21): 3904- 3920.
doi: 10.1016/j.molcel.2023.09.034 |
| 39 |
LIU K , WEI H , NONG W , et al. Nampt/SIRT2/LDHA pathway-mediated lactate production regulates follicular dysplasia in polycystic ovary syndrome[J]. Free Radic Biol Med, 2024, 225, 776- 793.
doi: 10.1016/j.freeradbiomed.2024.10.312 |
| 40 |
MATSUMOTO Y , NAKASHIMA T , CHO O , et al. Pyruvate-triggered TCA cycle regulation in Staphylococcus aureus promotes tolerance to betamethasone valerate[J]. Biochem Biophys Res Commun, 2020, 528 (2): 318- 321.
doi: 10.1016/j.bbrc.2020.05.035 |
| 41 |
YIEW N , FINCK B N . The mitochondrial pyruvate carrier at the crossroads of intermediary metabolism[J]. Am J Physiol Endocrinol Metab, 2022, 323 (1): E33- E52.
doi: 10.1152/ajpendo.00074.2022 |
| 42 |
GONZALES-FIGUEROA H , GONZALES-MOLFINO H M . Maturation of pig oocytes in vitro in a medium with pyruvate[J]. Braz J Med Biol Res, 2005, 38 (6): 869- 872.
doi: 10.1590/S0100-879X2005000600008 |
| 43 |
JOHNSON M T , FREEMAN E A , GARDNER D K , et al. Oxidative metabolism of pyruvate is required for meiotic maturation of murine oocytes in vivo[J]. Biol Reprod, 2007, 77 (1): 2- 8.
doi: 10.1095/biolreprod.106.059899 |
| [1] | ZHENG Yunchang, HOU Ruilin, LIANG Xiaohe, YANG Lidan, ZHANG Yinjiao, HUO Haonan, CHEN Weina, ZHANG Cui, LI Shijie. Analysis of Monoallelic Expression and DNA Methylation Status of Bovine FOXP2 Gene [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(9): 4369-4378. |
| [2] | MA Siqi, Lü Wenwen, CHEN Junzhen, LI Jianlin, LIU Yucheng, GUAN Tuan, DING Jian, LIU Haoran, YE Hongyan, YANG Li, FU Qiang, SHI Huijun. Construction of Recombinant Adenovirus with Bovine Enterovirus VP1 Gene and Evaluation of Its Immunogenicity in Mice [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(9): 4615-4625. |
| [3] | LIU Jiajin, WEN Xiaoqing, LUO Chunhai, JIA Hongdou, WANG Wei, LI Danyang, FU Shixin. The Influence of FoxO1 on Expression of Apoptotic Factors in Dairy Cow Endometrial Epithelial Cells Induced by High NEFA Levels [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(9): 4708-4717. |
| [4] | YANG Wenzhe, WANG Jinhao, ZHAO Zichen, ZHAO Tong, PAN Feilong, CHEN Fangfang, SHAO Wenqi, LIU Kexiang, ZHAO Shuchen, ZHAO Lijia. Analysis of the Impact of Curcumin on the Ferroptosis Pathway in Alleviating the Inflammatory Response Induced by LPS in Bovine Mammary Epithelial Cells [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(9): 4730-4740. |
| [5] | WAN Qiongfei, SHI Shanshan, GUO Ruonan, Lü Hang, HU Debao, GUO Yiwen, ZHANG Linlin, DING Xiangbin, GUO Hong, LI Xin. Screening and Functional Analysis of Key lncRNAs of Bovine Embryonic Muscle Development [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(8): 3802-3812. |
| [6] | ZHAO Tong, WANG Yahui, WU Tianyi, GAO Chen, GAO Xiaoxiao, ZHANG Lupei, GAO Huijiang, LI Junya. Effects of Overexpression and Interference of PRKD1 Gene on Osteogenic Differentiation of Bovine Osteoblasts [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(7): 3188-3198. |
| [7] | LIU Yuze, YU Zhuoya, GONG Zhiguo, REN Peipei, ZHAO Jiamin, MAO Wei, ZHANG Shuangyi, FENG Shuang. The Impact of Lipoprotein on the Secretion of Inflammatory Mediators and the Synthesis of Prostaglandin E2 in Bovine Bone Marrow-derived Macrophages Infected with Staphylococcus aureus [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(7): 3474-3483. |
| [8] | GAO Linna, JIANG Yingying, WANG Yue, SHI Qianqian, AN Zhenjiang, WANG Huili, SHEN Yangyang, CHEN Kunlin, ZHANG Leying. Construction of a Whole Genome Knockout Library of bMECs Based on CRISPR/Cas9 Technology [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(6): 2711-2723. |
| [9] | HAN Xitong, ZHANG Nan, ZHANG Ning, ZHANG Jiaxin. FLI Promotes in Vitro Maturation of Bovine Oocytes by Increasing the Glucose Metabolism Pathway [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(6): 2778-2789. |
| [10] | JIA Chaoying, ZHANG Huawei, LUO Xiuxin, LIU Qingyun, WANG Xiangru. Establishment of Mice Model Infected by Bovine Mannheimia haemolytica and the Immunogenicity of Inactivated Vaccine [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(5): 2312-2324. |
| [11] | ZHAO Ying, WANG Jinglei, WANG Meng, WANG Libin, ZHANG Qian, LI Zhijie, MA Xin, YU Sijiu, PAN Yangyang. Preparation and Characterization of Forsythiaside A and Kaempferol Encapsulated in Milk-derived Exosomes and Evaluation of Anti-inflammatory Effects in vitro [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(5): 2481-2495. |
| [12] | XIONG Keng, FAN Haojie, WANG Jie, ZHAO Shanjiang, ZHU Qingli, HU Zhihui, LUO Haoshu, ZHU Huabin. Recent Advances and Applications of Recombinant Follicle-Stimulating Hormone in Bovine Superovulation [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(5): 2047-2055. |
| [13] | ZHAO Wanyue, XU Xiaowen, CHANG Shushu, XIANG Zhijie, GUO Aizhen, CHEN Yingyu. Epidemiologic Investigation of the Major Viruses of the Bovine Respiratory Disease Complex [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(3): 1324-1335. |
| [14] | JIANG Huihua, ZHAO Long, GUO Kangkang. Effect of HE Gene Receptor Binding Domain Variation on Bovine Coronavirus Infection [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(3): 1336-1343. |
| [15] | ZHAO Wenyue, YANG Jing, SHAO Yilan, LI Jiaxuan, JIANG Yanping, CUI Wen, WANG Xiaona, TANG Lijie. Screening and Identification of Secretory Signal Peptide of Lactobacillus reuteri Expressing Lactoferrin Peptide [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(3): 1431-1440. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||