





Acta Veterinaria et Zootechnica Sinica ›› 2025, Vol. 56 ›› Issue (9): 4369-4378.doi: 10.11843/j.issn.0366-6964.2025.09.020
• Animal Genetics and Breeding • Previous Articles Next Articles
ZHENG Yunchang1( ), HOU Ruilin1(
), HOU Ruilin1( ), LIANG Xiaohe1, YANG Lidan1, ZHANG Yinjiao1, HUO Haonan1, CHEN Weina2, ZHANG Cui1, LI Shijie1,*(
), LIANG Xiaohe1, YANG Lidan1, ZHANG Yinjiao1, HUO Haonan1, CHEN Weina2, ZHANG Cui1, LI Shijie1,*( )
)
Received:2025-02-19
Online:2025-09-23
Published:2025-09-30
Contact:
LI Shijie
E-mail:zyc15831601852@163.com;13780586712@163.com;lishijie20005@163.com
CLC Number:
ZHENG Yunchang, HOU Ruilin, LIANG Xiaohe, YANG Lidan, ZHANG Yinjiao, HUO Haonan, CHEN Weina, ZHANG Cui, LI Shijie. Analysis of Monoallelic Expression and DNA Methylation Status of Bovine FOXP2 Gene[J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(9): 4369-4378.
Table 1
Primers information"
| 引物名称 Primer name | 引物序列(5′→3′) Primer sequence | 用途 Application | 扩增片段/bp Amplicon size | 退火温度/℃ Tm | PCR类型 Type of PCR |
| q-F | CAGTACGTCATAATCTTAGCCT | 表达量的检测 | 316 | 60 | cDNA |
| q-R | CCATTGCTGTCGATATGATCC | ||||
| GA-F | AACCACGAGAAGTATAAC | 内参基因 | 223 | 60 | cDNA |
| GA-R | AAGCAGGGATGATATTC | ||||
| g-F | CCAATCCTTCCCTCCTCAA | 鉴定杂合子 | 659 | 52 | DNA |
| g-R | TAATGAAACTAACACTATTTTTGG | ||||
| c-F | GACAGCAATGGGAACAG | 等位基因分析 | 619 | 53 | cDNA |
| c-R | CATTTGCTTTCTACAGGATG | ||||
| FO20-OF-1 | GGTTGGGTTTGTATTGTGGA | 甲基化分析 | 458 | 52 | |
| FO20-OR-1 | TTTCACTCTATACACAAAAACAC | ||||
| FO20-IF-1 | GTTGGAAGGGAGGTTATAGT | 372 | 53 | ||
| FO20-IR-1 | TCCTATAAATACAAACAACCAAAC | ||||
| FOXP2-OF | GTGTGTGTTTGTGTATTAGT | 595 | 52 | ||
| FOXP2-OR | AAAAAAAAAAAACTACCAAAACTACC | ||||
| FOXP2-IF | TTAAAGGTAGAAGTTGGTGTTGT | 270 | 53 | ||
| FOXP2-IR1 | ACTACCAACCCCTCCCCTATC | ||||
| FO249-OF-1 | GGTGTGTTGTTTGTGGGTTT | 478 | 56 | ||
| FO249-OR-1 | ACACATCTACTATTTCCACACACA | ||||
| FO249-IF-1 | GTAAGTTTGTTTATTTTTATTTTGG | 293 | 53 | ||
| FO249-IR-2 | AAACAACCTCCCCTCTCTAA |

Fig. 1
The splice structure of the bovine FOXP2 gene and the location of primers The bovine FOXP2 gene structure of the mRNA and 8 predicted splice variants (X1-X8) sequences. Green, yellow and red arrows indicate primers used for analysis of gene expression levels, SNP identification and allelic expression. The SNP of g.53771685G > A on the last exon is located in the common sequence of all FOXP2 splices"
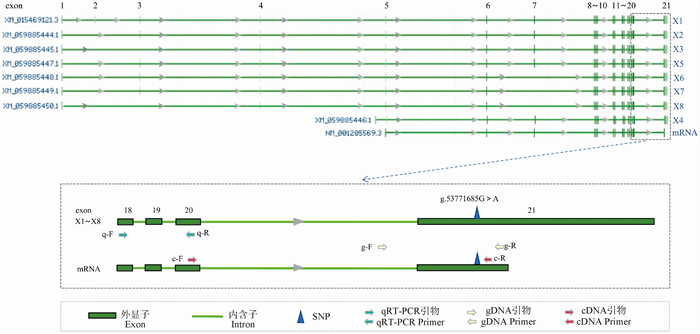
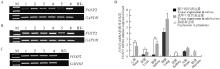
Fig. 2
The expression of FOXP2 gene in bovine tissues and placentas A. The expression of bovine FOXP2 gene in calf tissues. B. The expression of bovine FOXP2 gene in adult bovine tissues: 1-6. Heart, liver, spleen, lung, kidney and brain. C. The expression of bovine FOXP2 gene in bovine placenta: 1-4. RT-PCR products obtained from different placentas; M. DL2000 DNA Marker; RT-. Negative control. D. The expression levels of bovine FOXP2 gene in tissues and placentas: *. P < 0.05 represents significant difference; **. P < 0.01 represents extremely significant difference"
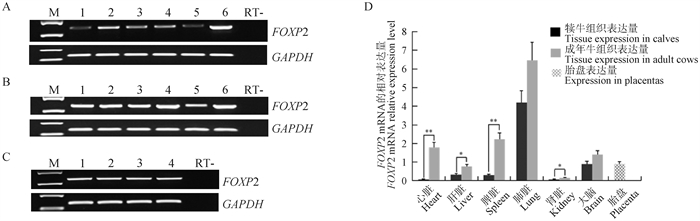

Fig. 3
The allelic expression analysis of FOXP2 gene in bovine tissues and placentas A. The allelic expression of FOXP2 gene in calf tissues; B. The allelic expression of FOXP2 gene in adult bovine tissues; C. Imprinting analysis of FOXP2 gene in placentas. The SNP is indicated with the dashed box"
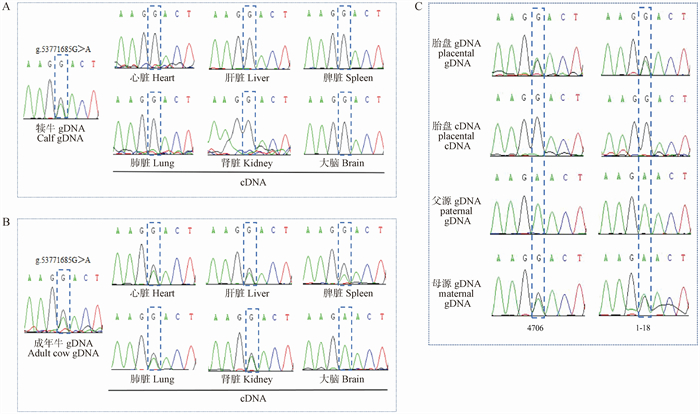

Fig. 4
The location of the CpG island of the bovine FOXP2 gene and the methylation status analysis A. The location of CpG20 and CpG249 and the 3 detected regions of bovine FOXP2 gene. B. Methylation status in the 3 regions tested. Only the methylation status of placenta, calf heart, adult bovine heart, adult bovine brain and spermatozoa are showed. The white circles are the non-methylated status, the black circles are the methylation status"

| 1 |
MACDONALD W A , MANN M R W . Long noncoding RNA functionality in imprinted domain regulation[J]. PLoS Genet, 2020, 16 (8): e1008930.
doi: 10.1371/journal.pgen.1008930 |
| 2 |
THAMBAN T , AGARWAAL V , KHOSLA S . Role of genomic imprinting in mammalian development[J]. J Biosci, 2020, 45, 20.
doi: 10.1007/s12038-019-9984-1 |
| 3 |
BUTLER M G . Imprinting disorders in humans: a review[J]. Curr Opin Pediatr, 2020, 32 (6): 719- 729.
doi: 10.1097/MOP.0000000000000965 |
| 4 |
ELBRACHT M , MACKAY D , BEGEMANN M , et al. Disturbed genomic imprinting and its relevance for human reproduction: causes and clinical consequences[J]. Hum Reprod Update, 2020, 26 (2): 197- 213.
doi: 10.1093/humupd/dmz045 |
| 5 |
WEINBERG-SHUKORN A , BEN-YAIR R , TAKAHASHI N , et al. Balanced gene dosage control rather than parental origin underpins genomic imprinting[J]. Nat Commun, 2022, 13 (1): 4391.
doi: 10.1038/s41467-022-32144-z |
| 6 |
GLASER J , IRANZO J , BORENSZTEIN M , et al. The imprinted Zdbf2 gene finely tunes control of feeding and growth in neonates[J]. Elife, 2022, 11, e65641.
doi: 10.7554/eLife.65641 |
| 7 |
SAPEHIA D , MAHAJAN A , SINGH P , et al. Enrichment of trimethyl histone 3 lysine 4 in the Dlk1 and Grb10 genes affects pregnancy outcomes due to dietary manipulation of excess folic acid and low vitamin B12[J]. Biol Res, 2024, 57 (1): 85.
doi: 10.1186/s40659-024-00557-3 |
| 8 |
ANGIOLINI E , SANDOVICI I , COAN P M , et al. Deletion of the imprinted Phlda2 gene increases placental passive permeability in the mouse[J]. Genes (Basel), 2021, 12 (5): 639.
doi: 10.3390/genes12050639 |
| 9 |
LIU S , YU Y , ZHANG S , et al. Epigenomics and genotype-phenotype association analyses reveal conserved genetic architecture of complex traits in cattle and human[J]. BMC Biol, 2020, 18 (1): 80.
doi: 10.1186/s12915-020-00792-6 |
| 10 | 王丁香, 赵红波. 牛印记基因的研究进展[J]. 中国牛业科学, 2022, 48 (6): 80- 84. |
| WANG D X , ZHAO H B . Research advances on imprinted genes in cattle[J]. China Cattle Science, 2022, 48 (6): 80- 84. | |
| 11 |
BENVENUTO M , PALUMBO P , DI MURO E , et al. Identification of a novel FOXP1 variant in a patient with hypotonia, intellectual disability, and severe speech impairment[J]. Genes (Basel), 2023, 14 (10): 1958.
doi: 10.3390/genes14101958 |
| 12 |
PERUMAL C M , THULO M , BUTHELEZI S , et al. Unraveling the interplay between the leucine zipper and forkhead domains of FOXP2: Implications for DNA binding, stability and dynamics[J]. Proteins, 2024, 92 (10): 1177- 1189.
doi: 10.1002/prot.26699 |
| 13 |
CHE F , LI C , ZHANG L , et al. Novel FOXP2 variant associated with speech and language dysfunction in a Chinese family and literature review[J]. J Appl Genet, 2024, 65 (2): 367- 373.
doi: 10.1007/s13353-024-00849-0 |
| 14 | TURNER S J , HILDEBRAND M S , BLOCK S , et al. Small intragenic deletion in FOXP2 associated with childhood apraxia of speech and dysarthria[J]. Am J Med Genet A, 2013, 161A (9): 2321- 2326. |
| 15 |
SU W , HU S , ZHOU L , et al. FOXP2 inhibits the aggressiveness of lung cancer cells by blocking TGFβ signaling[J]. Oncol Lett, 2024, 27 (5): 227.
doi: 10.3892/ol.2024.14361 |
| 16 |
YANG F , XIAO Z , ZHANG S . FOXP2 regulates thyroid cancer cell proliferation and apoptosis via transcriptional activation of RPS6KA6[J]. Exp Ther Med, 2022, 23 (6): 434.
doi: 10.3892/etm.2022.11361 |
| 17 |
THOMAS A C , FROST J M , ISHIDA M , et al. The speech gene FOXP2 is not imprinted[J]. J Med Genet, 2012, 49 (11): 669- 670.
doi: 10.1136/jmedgenet-2012-101242 |
| 18 |
ADEGBOLA A A , COX G F , BRADSHAW E M , et al. Monoallelic expression of the human FOXP2 speech gene[J]. Proc Natl Acad Sci U S A, 2015, 112 (22): 6848- 6854.
doi: 10.1073/pnas.1411270111 |
| 19 |
REGMI S , GIHA L , ALI A , et al. Methylation is maintained specifically at imprinting control regions but not other DMRs associated with imprinted genes in mice bearing a mutation in the Dnmt1 intrinsically disordered domain[J]. Front Cell Dev Biol, 2023, 11, 1192789.
doi: 10.3389/fcell.2023.1192789 |
| 20 |
BARLOW D P , BARTOLOMEI M S . Genomic imprinting in mammals[J]. Cold Spring Harb Perspect Biol, 2014, 6 (2): a018382.
doi: 10.1101/cshperspect.a018382 |
| 21 |
MAS-PARES B , CARRERAS-BADOSA G , GOMEZ-VILARRUBLA A , et al. Sex dimorphic associations of Prader-Willi imprinted gene expressions in umbilical cord with prenatal and postnatal growth in healthy infants[J]. World J Pediatr, 2025, 21 (1): 100- 112.
doi: 10.1007/s12519-024-00865-4 |
| 22 |
KANG J , LI Q , LIU J , et al. Exploring the cellular and molecular basis of murine cardiac development through spatiotemporal transcriptome sequencing[J]. Gigascience, 2025, 14, giaf012.
doi: 10.1093/gigascience/giaf012 |
| 23 |
WEINBERG-SHUKRON A , YOUNGSON N A , FERGUSON-SMITH A C , et al. Epigenetic control and genomic imprinting dynamics of the Dlk1-Dio3 domain[J]. Front Cell Dev Biol, 2023, 11, 1328806.
doi: 10.3389/fcell.2023.1328806 |
| 24 |
CROCCO P , DE RANGO F , BRUNO F , et al. Genetic variability of FOXP2 and its targets CNTNAP2 and PRNP in frontotemporal dementia: A pilot study in a southern Italian population[J]. Heliyon, 2024, 10 (11): e31624.
doi: 10.1016/j.heliyon.2024.e31624 |
| 25 |
YIN M , YU W , LI W , et al. DNA methylation and gene expression changes in mouse pre- and post-implantation embryos generated by intracytoplasmic sperm injection with artificial oocyte activation[J]. Reprod Biol Endocrinol, 2021, 19 (1): 163.
doi: 10.1186/s12958-021-00845-7 |
| 26 |
ZHANG Y , ZHANG C , CHEN W , et al. The landscape of allelic expression and DNA methylation at the bovine SGCE/PEG10 locus[J]. Anim Genet, 2024, 55 (3): 452- 456.
doi: 10.1111/age.13429 |
| 27 | SCHUFF M , STRONG A D , WELBORN L K , et al. Imprinting as basis for complex evolutionary novelties in Eutherians[J]. Biology (Basel), 2024, 13 (9): 682. |
| 28 |
ISLES A R . The contribution of imprinted genes to neurodevelopmental and neuropsychiatric disorders[J]. Transl Psychiatry, 2022, 12 (1): 210.
doi: 10.1038/s41398-022-01972-4 |
| 29 |
CINDROVA-DAVIES T , SFERRUZZI-PERRI A N . Human placental development and function[J]. Semin Cell Dev Biol, 2022, 131, 66- 77.
doi: 10.1016/j.semcdb.2022.03.039 |
| 30 |
HARA S , MATSUHISA F , KITAJIMA S , et al. Identification of responsible sequences which mutations cause maternal H19-ICR hypermethylation with Beckwith-Wiedemann syndrome-like overgrowth[J]. Commun Biol, 2024, 7 (1): 1605.
doi: 10.1038/s42003-024-07323-x |
| 31 | DASKEVICIUTE D , CHAPPELL-MAOR L , SAINTY B , et al. Non-canonical imprinting, manifesting as post-fertilization placenta-specific parent-of-origin dependent methylation, is not conserved in humans[J]. Hum Mol Genet, 2025, 17, ddaf009. |
| 32 |
RICHARD ALBER J , KOBAYASHI T , INOUE A , et al. Conservation and divergence of canonical and non-canonical imprinting in murids[J]. Genome Biol, 2023, 24 (1): 48.
doi: 10.1186/s13059-023-02869-1 |
| 33 |
KANEKO-ISHINO T , ISHINO F . The evolutionary advantage in mammals of the complementary monoallelic expression mechanism of genomic imprinting and its emergence from a defense against the insertion into the host genome[J]. Front Genet, 2022, 13, 832983.
doi: 10.3389/fgene.2022.832983 |
| 34 |
DENG Q , DU Y , WANG Z , et al. Identification and validation of a DNA methylation driven gene based prognostic model for clear cell renal cell carcinoma[J]. BMC Genomics, 2023, 24 (1): 307.
doi: 10.1186/s12864-023-09416-z |
| 35 |
BRENET F , MOH M , FUNK P , et al. DNA methylation of the first exon is tightly linked to transcriptional silencing[J]. PLoS One, 2011, 6 (1): e14524.
doi: 10.1371/journal.pone.0014524 |
| 36 |
刘晓倩, 靳兰杰, 董艳秋, 等. DNA甲基化调控牛AQP1基因的胎盘特异性印记[J]. 畜牧兽医学报, 2021, 52 (8): 2181- 2189.
doi: 10.11843/j.issn.0366-6964.2021.08.011 |
|
LIU X Q , JIN L J , DONG Y Q , et al. DNA methylation regulate the genomic imprinting of AQP1 gene specific in bovine placenta[J]. Acta Veterinaria et Zootechnica Sinica, 2021, 52 (8): 2181- 2189.
doi: 10.11843/j.issn.0366-6964.2021.08.011 |
|
| 37 |
CHEN Z , ZHANG Y . Maternal H3K27me3-dependent autosomal and X chromosome imprinting[J]. Nat Rev Genet, 2020, 21 (9): 555- 571.
doi: 10.1038/s41576-020-0245-9 |
| 38 |
HANNA C W , KELSEY G . Features and mechanisms of canonical and noncanonical genomic imprinting[J]. Genes Dev, 2021, 35 (11-12): 821- 834.
doi: 10.1101/gad.348422.121 |
| [1] | LIN Xiao, LI Ruijie, LIU Long, GENG Tuoyu, GONG Daoqing. Research Progress on Sex Determining Genes and Their Methylation Regulation in Animals [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(9): 4129-4142. |
| [2] | WANG Rui, HENG Nuo, HU Yingfan, WANG Huan, ZHU Ni, HE Wei, XUAN Xiuli, HU Zhihui, XIONG Keng, GONG Jianfei, HAO Haisheng, ZHU Huabin, ZHAO Shanjiang. Mechanisms of Developmental Competence Differences of Oocytes from Different Grades of Bovine Cumulus-Oocyte Complexes after in Vitro Maturation [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(9): 4432-4451. |
| [3] | MA Siqi, Lü Wenwen, CHEN Junzhen, LI Jianlin, LIU Yucheng, GUAN Tuan, DING Jian, LIU Haoran, YE Hongyan, YANG Li, FU Qiang, SHI Huijun. Construction of Recombinant Adenovirus with Bovine Enterovirus VP1 Gene and Evaluation of Its Immunogenicity in Mice [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(9): 4615-4625. |
| [4] | LIU Jiajin, WEN Xiaoqing, LUO Chunhai, JIA Hongdou, WANG Wei, LI Danyang, FU Shixin. The Influence of FoxO1 on Expression of Apoptotic Factors in Dairy Cow Endometrial Epithelial Cells Induced by High NEFA Levels [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(9): 4708-4717. |
| [5] | YANG Wenzhe, WANG Jinhao, ZHAO Zichen, ZHAO Tong, PAN Feilong, CHEN Fangfang, SHAO Wenqi, LIU Kexiang, ZHAO Shuchen, ZHAO Lijia. Analysis of the Impact of Curcumin on the Ferroptosis Pathway in Alleviating the Inflammatory Response Induced by LPS in Bovine Mammary Epithelial Cells [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(9): 4730-4740. |
| [6] | WAN Qiongfei, SHI Shanshan, GUO Ruonan, Lü Hang, HU Debao, GUO Yiwen, ZHANG Linlin, DING Xiangbin, GUO Hong, LI Xin. Screening and Functional Analysis of Key lncRNAs of Bovine Embryonic Muscle Development [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(8): 3802-3812. |
| [7] | ZHAO Tong, WANG Yahui, WU Tianyi, GAO Chen, GAO Xiaoxiao, ZHANG Lupei, GAO Huijiang, LI Junya. Effects of Overexpression and Interference of PRKD1 Gene on Osteogenic Differentiation of Bovine Osteoblasts [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(7): 3188-3198. |
| [8] | LIU Yuze, YU Zhuoya, GONG Zhiguo, REN Peipei, ZHAO Jiamin, MAO Wei, ZHANG Shuangyi, FENG Shuang. The Impact of Lipoprotein on the Secretion of Inflammatory Mediators and the Synthesis of Prostaglandin E2 in Bovine Bone Marrow-derived Macrophages Infected with Staphylococcus aureus [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(7): 3474-3483. |
| [9] | GAO Linna, JIANG Yingying, WANG Yue, SHI Qianqian, AN Zhenjiang, WANG Huili, SHEN Yangyang, CHEN Kunlin, ZHANG Leying. Construction of a Whole Genome Knockout Library of bMECs Based on CRISPR/Cas9 Technology [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(6): 2711-2723. |
| [10] | HAN Xitong, ZHANG Nan, ZHANG Ning, ZHANG Jiaxin. FLI Promotes in Vitro Maturation of Bovine Oocytes by Increasing the Glucose Metabolism Pathway [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(6): 2778-2789. |
| [11] | JIA Chaoying, ZHANG Huawei, LUO Xiuxin, LIU Qingyun, WANG Xiangru. Establishment of Mice Model Infected by Bovine Mannheimia haemolytica and the Immunogenicity of Inactivated Vaccine [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(5): 2312-2324. |
| [12] | ZHAO Ying, WANG Jinglei, WANG Meng, WANG Libin, ZHANG Qian, LI Zhijie, MA Xin, YU Sijiu, PAN Yangyang. Preparation and Characterization of Forsythiaside A and Kaempferol Encapsulated in Milk-derived Exosomes and Evaluation of Anti-inflammatory Effects in vitro [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(5): 2481-2495. |
| [13] | XIONG Keng, FAN Haojie, WANG Jie, ZHAO Shanjiang, ZHU Qingli, HU Zhihui, LUO Haoshu, ZHU Huabin. Recent Advances and Applications of Recombinant Follicle-Stimulating Hormone in Bovine Superovulation [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(5): 2047-2055. |
| [14] | ZHAO Wanyue, XU Xiaowen, CHANG Shushu, XIANG Zhijie, GUO Aizhen, CHEN Yingyu. Epidemiologic Investigation of the Major Viruses of the Bovine Respiratory Disease Complex [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(3): 1324-1335. |
| [15] | JIANG Huihua, ZHAO Long, GUO Kangkang. Effect of HE Gene Receptor Binding Domain Variation on Bovine Coronavirus Infection [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(3): 1336-1343. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||