





Acta Veterinaria et Zootechnica Sinica ›› 2025, Vol. 56 ›› Issue (6): 2711-2723.doi: 10.11843/j.issn.0366-6964.2025.06.016
• Animal Genetics and Breeding • Previous Articles Next Articles
GAO Linna1,2( ), JIANG Yingying2,3, WANG Yue2,3, SHI Qianqian1,2, AN Zhenjiang2,3, WANG Huili2, SHEN Yangyang2, CHEN Kunlin2,*(
), JIANG Yingying2,3, WANG Yue2,3, SHI Qianqian1,2, AN Zhenjiang2,3, WANG Huili2, SHEN Yangyang2, CHEN Kunlin2,*( ), ZHANG Leying1,*(
), ZHANG Leying1,*( )
)
Received:2024-11-19
Online:2025-06-23
Published:2025-06-25
Contact:
CHEN Kunlin, ZHANG Leying
E-mail:13292751837@163.com;chenkunlin@jaas.ac.cn;zhangly056000@126.com
CLC Number:
GAO Linna, JIANG Yingying, WANG Yue, SHI Qianqian, AN Zhenjiang, WANG Huili, SHEN Yangyang, CHEN Kunlin, ZHANG Leying. Construction of a Whole Genome Knockout Library of bMECs Based on CRISPR/Cas9 Technology[J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(6): 2711-2723.

Fig. 2
Distribution map of sgRNA in plasmid library A. Histgram of gRNA sequencing depth, horizontal coordinate represents the log2 (sequencing depth), vertical coordinate represents the number of gRNAs; B. Skew ration of gRNA distribution, horizontal coordinate represents the sequencing depth, vertical coordinate represents the frequency of gRNA accumulation, lower-left value: reads ≤359 for 10% sgRNAs in sgRNAs, middle-left value: the number of reads ≤842 for 90% sgRNAs, upper-left value : the ratio of the two skew ration is 2.35"
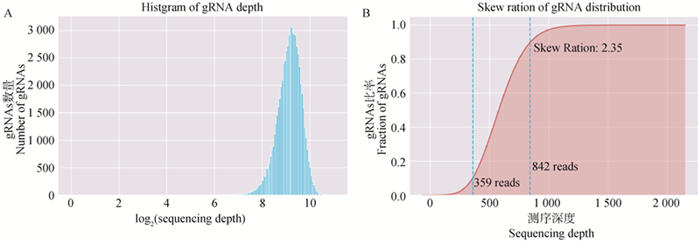
Table 6
The measurement results of the GFP reference virus and library lentivirus titer"
| 样品名称 Sample | 感染时细胞数目 Cell number | 病毒使用量/mL Virus usage amount | 病毒滴度/(TU·mL-1) Virus titer |
| 文库慢病毒Library lentivirus | 1.60×105 | 5.00×10-5 | 7.30×108 |
| GFP参照病毒 GFP reference virus | 1.60×105 | 5.00×10-5 | 2.61×107 |
| 文库实际滴度 Actual titer of the library | / | / | 7.01×108 |

Fig. 7
Cell library quality control chart A. Quality scores across all bases, horizontal coordinate represents the base position of reads, vertical coordinate represents the quality score distribution of that base position, blue line represents the average value, green area represents the quality score of Q28 and above, the base accuracy is very high, yellow area represents the quality score between Q20-Q28, the base accuracy is medium, red area represents the quality score below Q20, the base uncertainty is high; B. Quality score distribution over all sequences, the horizontal coordinate represents the average quality score of the reads, the vertical coordinate is the number of reads under the specific score"
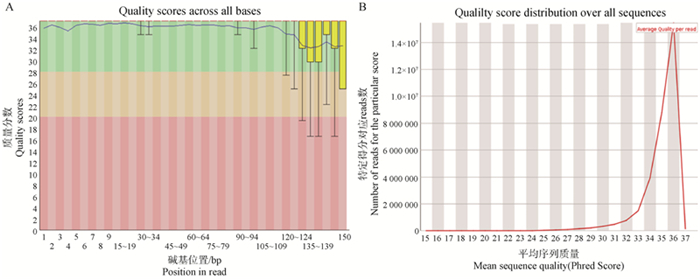
Table 8
Top 20 sgRNAs in cell library by count number"
| 基因名称Gene name | 向导RNA(5′→3′)sgRNA | sgRNA的count数Count reads |
| GC | TTTGTTGGCTCTACGTAAGT | 1 376 398 |
| PALB2 | TTTGTTGGCCGCCGGTCAGA | 727 513 |
| GUCY1A2 | TTTGTTGGAATGGTCTGCAT | 631 165 |
| CELF3 | TTTGTTGAGGAGAGCGCGGC | 574 104 |
| C1QTNF7 | TTTGTTCTGGCTGGCGCTAG | 451 616 |
| POLR3H | TTTGTTCAACTCTTCGGCGA | 398 934 |
| ORMDL1 | TTTGTTAAGGTCCAAGCAAC | 372 299 |
| LOC787250 | TTTGTGTTTGAGGTCGAGCT | 335 331 |
| NC-sgRNA-1698 | TTTGTGTGGGTAGGTCCGGG | 281 296 |
| LOC100336208 | TTTGTGTCTGCTACTGTCAT | 279 009 |
| SCN2B | TTTGTGGTTCACGGTGTAGC | 275 791 |
| PRPF38A | TTTGTGGGTGGTGTCTACGG | 275 601 |
| EDAR | TTTGTGGGCGAGCTGTGGCG | 272 757 |
| CDHR1 | TTTGTGGGCACGCCCTACTA | 271 241 |
| LOC112447450 | TTTGTGGCTCTCATGGTTAT | 252 815 |
| DYSF_1 | TTTGTGGATCCGCGTCCGCT | 235 587 |
| TAFA1_2 | TTTGTGGATAAGTGCTTGCG | 235 579 |
| SNX4_3 | TTTGTGGAAAGACGACGAAT | 233 111 |
| BRINP1_1 | TTTGTGGAAAGACACCGTCA | 227 912 |
| FANCB_2 | TTTGTGCGTACTCTCTTGAA | 213 775 |
| 1 |
KHADEMPAR S , FAMILGHADAKCHI S , MOTLAGH R A , et al. CRISPR-Cas9 in genome editing: Its function and medical applications[J]. J Cell Physiol, 2019, 234 (5): 5751- 5761.
doi: 10.1002/jcp.27476 |
| 2 |
ZHANG B . CRISPR/Cas gene therapy[J]. J Cell Physiol, 2021, 236 (4): 2459- 2481.
doi: 10.1002/jcp.30064 |
| 3 |
JANIK E , NIEMCEWICZ M , CEREMUGA M , et al. Various aspects of a gene editing system-CRISPR-Cas9[J]. Int J Mol Sci, 2020, 21 (24): e9604.
doi: 10.3390/ijms21249604 |
| 4 |
HORODECKA K , DVCHLER M . CRISPR/Cas9: Principle, applications, and delivery through extracellular vesicles[J]. Int J Mol Sci, 2021, 22 (11): 6072.
doi: 10.3390/ijms22116072 |
| 5 |
BHATTACHARYA S , SATPATI P . Insights into the mechanism of CRISPR/Cas9-based genome editing from molecular dynamics simulations[J]. ACS Omega, 2023, 8 (2): 1817- 1837.
doi: 10.1021/acsomega.2c05583 |
| 6 |
WANG S W , GAO C , ZHENG Y M , et al. Current applications and future perspective of CRISPR/Cas9 gene editing in cancer[J]. Mol Cancer, 2022, 21 (1): 1- 27.
doi: 10.1186/s12943-021-01470-z |
| 7 |
FU Y W , DAI X Y , WANG W T , et al. Dynamics and competition of CRISPR-Cas9 ribonucleoproteins and AAV donor-mediated NHEJ, MMEJ and HDR editing[J]. Nucleic Acids Res, 2021, 49 (2): 969- 985.
doi: 10.1093/nar/gkaa1251 |
| 8 | 杨丽芸, 陈丽娇, 李善刚. CRISPR/Cas9系统诱导DNA断裂的修复机制研究进展[J]. 中国细胞生物学学报, 2022, 44 (03): 500- 511. |
| YANG L Y , CHEN L J , LI S G . Research progress on repair mechanism of DNA breakage induced by CRISPR/Cas9 System[J]. Chinese Journal of Cell Biology, 2022, 44 (3): 500- 511. | |
| 9 |
SHALEM O , SANJANA N E , HARTENIAN E , et al. Genome-scale CRISPR-Cas9 knockout screening in human cells[J]. Science, 2014, 343 (6166): 84- 87.
doi: 10.1126/science.1247005 |
| 10 |
YU J S L , YUSA K . Genome-wide CRISPR-Cas9 screening in mammalian cells[J]. Methods, 2019, 164-165, 29- 35.
doi: 10.1016/j.ymeth.2019.04.015 |
| 11 |
HAN J , PEREZ J T , CHEN C , et al. Genome-wide CRISPR/Cas9 screen identifies host factors essential for influenza virus replication[J]. Cell Rep, 2018, 23 (2): 596- 607.
doi: 10.1016/j.celrep.2018.03.045 |
| 12 |
YI C , CAI C , CHENG Z , et al. Genome-wide CRISPR-Cas9 screening identifies the CYTH2 host gene as a potential therapeutic target of influenza viral infection[J]. Cell Rep, 2022, 38 (13): 110559.
doi: 10.1016/j.celrep.2022.110559 |
| 13 |
SHUE B , CHIRAMEL A I , CERIKAN B , et al. Genome-Wide CRISPR screen identifies RACK1 as a critical host factor for flavivirus replication[J]. J Virol, 2021, 95 (24): e0059621.
doi: 10.1128/JVI.00596-21 |
| 14 |
KIM G , NAKAYAMA L , BLUM J A , et al. Genome-wide CRISPR screen reveals v-ATPase as a drug target to lower levels of ALS protein ataxin-2[J]. Cell Rep, 2022, 41 (4): 111508.
doi: 10.1016/j.celrep.2022.111508 |
| 15 | 聂震宇. 全基因组文库筛选膀胱癌吡柔比星耐药基因AKR1C1的机制研究[D]. 长沙: 中南大学, 2023. |
| NIE Z Y. Genome-wide sereening identifies genes AKR1C1 critical for resistance to pirarubicin in bladder cancer[D]. Changsha: Central South University, 2023. (in Chinese) | |
| 16 |
HU J , GUAN X , ZHAO M , et al. Genome-wide CRISPR-Cas9 knockout screening reveals a TSPAN3-mediated endo-lysosome pathway regulating the degradation of α-synuclein oligomers[J]. Mol Neurobiol, 2023, 60 (11): 6731- 6747.
doi: 10.1007/s12035-023-03495-5 |
| 17 | 潘冰心. 利用CRISPR/Cas9全基因组文库筛选HCT 116细胞增殖和辐射相关基因[D]. 合肥: 安徽医科大学, 2019. |
| PAN B X. Genome-Scale CRISPR/Cas9 screening for proliferation and radiation-related genes in HCT 116 cells[D]. Hefei: Anhui Medical University, 2019. (in Chinese) | |
| 18 |
MARTINEZ S , WU S , GEUENICH M , et al. In vivo CRISPR screens reveal SCAF1 and USP15 as drivers of pancreatic cancer[J]. Nat Commun, 2024, 15 (1): 1- 15.
doi: 10.1038/s41467-023-43650-z |
| 19 |
WANG S , XIONG Y , LUO Y , et al. Genome-wide CRISPR screens identify PKMYT1 as a therapeutic target in pancreatic ductal adenocarcinoma[J]. EMBO Mol Med, 2024, 16 (5): 1115- 1142.
doi: 10.1038/s44321-024-00060-y |
| 20 | PENG R , CAO J , ZHANG C , et al. In vivo CRISPR screen identifies LTN1 as a novel tumor suppressor ubiquitinating insulin-like growth factor 2 mRNA-binding protein 1 in hepatocellular carcinoma[J]. Hepatol Commun, 2023, 7 (10): e0256. |
| 21 |
TUANO N K , BEESLEY J , MANNING M , et al. CRISPR screens identify gene targets at breast cancer risk loci[J]. Genome Biol, 2023, 24 (1): 1- 23.
doi: 10.1186/s13059-022-02832-6 |
| 22 |
DAS T , ANAND U , PAL T , et al. Exploring the potential of CRISPR/Cas genome editing for vegetable crop improvement: An overview of challenges and approaches[J]. Biotechnol Bioeng, 2023, 120 (5): 1215- 1228.
doi: 10.1002/bit.28344 |
| 23 | 殷文晶, 陈振概, 黄佳慧, 等. 基于CRISPR-Cas9基因编辑技术在作物中的应用[J]. 生物工程学报, 2023, 39 (2): 399- 424. |
| YIN W J , CHEN Z G , HUANG J H , et al. Application of CRISPR-Cas9 gene editing technology in crop breeding[J]. Chinese Journal of Biotechnology, 2023, 39 (2): 399- 424. | |
| 24 | 李可, 吴传银, 隋毅. CRISPR/Cas基因编辑技术在水稻育种中的研究进展[J]. 科学通报, 2025, 1- 12. |
| LI K , WU C Y , SUI Y . Research progress of CRISPR/Cas gene editing technology in rice breeding[J]. Chinese Science Bulletin, 2025, 1- 12. | |
| 25 |
鲍艳春, 戴伶俐, 刘在霞, 等. CRISPR/Cas9系统在畜禽遗传改良中研究进展[J]. 遗传, 2024, 46 (3): 219- 231.
doi: 10.3760/cma.j.cn231536-20231228-00079-1 |
|
BAO Y C , DAI L L , LIU Z X , et al. Progress on CRISPR/Cas9 system in the genetic improvement of livestock and poultry[J]. Hereditas, 2024, 46 (3): 219- 231.
doi: 10.3760/cma.j.cn231536-20231228-00079-1 |
|
| 26 | 常珈菘. 家蚕全基因组编辑细胞库的构建及其应用[D]. 重庆: 西南大学, 2020. |
| CHANG J S. Genome-wide CRISPR screening in bombyx mori cells[D]. Chongqing: Southwest University, 2020. (in Chinese) | |
| 27 | 赵长志. 猪全基因组CRISPR/Cas9敲除文库的构建及筛选病毒抗性关键宿主因子[D]. 武汉: 华中农业大学, 2019. |
| ZHAO C Z. Construction of pig genome-scale CRISPR/Cas9 knockout library and screening of key host factors for virus resistance[D]. Wuhan: Huazhong Agricultural University, 2019. (in Chinese) | |
| 28 | 徐娟, 刘忠媛, 刘彦峰, 等. 基于CRISPR-Cas9技术的鸡成纤维细胞系全基因组敲除文库的建立与初步应用[J]. 中国动物传染病学报, 2022, 30 (5): 50- 59. |
| XU J , LIU Z Y , LIU Y F , et al. Development and preliminary application of chicken fibroblast cell line based on genome-scale CRISPR-Cas9 knockout screening[J]. Chinese Journal of Animal Infectious Diseases, 2022, 30 (5): 50- 59. | |
| 29 | 李晓娇. 鸡全基因组CRISPR高通量筛选技术的建立与应用[D]. 广州: 仲恺农业工程学院, 2023. |
| LI X J. Establishment and application of chicken genome-wide CRISPR high-throughput screening technology[D]. Guangzhou: Zhongkai University of Agricultural and Engineering, 2023. (in Chinese) | |
| 30 | 李岚. 利用CRISPR文库鉴定绒山羊毛乳头细胞增殖的必需基因研究[D]. 杨凌: 西北农林科技大学, 2021. |
| LI L. Screening essential genes for the proliferation of cashmere goat dermal papilla cells by CRISPR library[D]. Yangling: Northwest A&F University, 2021. (in Chinese) | |
| 31 |
丁修虎, 林志平, 赵芳, 等. 利用CRISPR/Cas9技术制备BLG基因敲除牛乳腺上皮细胞系[J]. 畜牧兽医学报, 2024, 55 (10): 4475- 4488.
doi: 10.11843/j.issn.0366-6964.2024.10.020 |
|
DING X H , LIN Z P , ZHAO F , et al. Highly efficient BLG knockout in cow mammary epithelial cells by using CRISPR/Cas9[J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55 (10): 4475- 4488.
doi: 10.11843/j.issn.0366-6964.2024.10.020 |
|
| 32 | 李艳, 卞志标, 翟少伦, 等. 猪流行性腹泻病毒锁核酸探针荧光定量PCR检测方法的建立[J]. 广东畜牧兽医科技, 2024, 49 (2): 30- 36. |
| LI Y , BIAN Z B , ZHAI S L , et al. Methodology development of a locked nucleic acid-based fluorescent quantitation RT-PCR for detecting porcine epidemic diarrhea virus[J]. Guangdong Journal of Animal And Veterinary Science, 2024, 49 (2): 30- 36. | |
| 33 | 赵娅娅, 袁利明, 华进联. 基因编辑技术在猪分子育种中的研究进展及发展趋势[J]. 农业生物技术学报, 2024, 32 (8): 1939- 1948. |
| ZHAO Y Y , YUAN L M , HUA J L . Research progress and development trend of gene editing technology in pig (sus scrofa) molecular breeding[J]. Journal of Agricultural Biotechnology, 2024, 32 (8): 1939- 1948. | |
| 34 |
ZHOU Y , ZHU S , CAI C , et al. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells[J]. Nature, 2014, 509 (7501): 487- 491.
doi: 10.1038/nature13166 |
| 35 |
ZHANG M L , LI H B , JIN Y . Application and perspective of CRISPR/Cas9 genome editing technology in human diseases modeling and gene therapy[J]. Front Genet, 2024, 15, 1364742.
doi: 10.3389/fgene.2024.1364742 |
| 36 |
CHEN S , SANJANA N E , ZHENG K , et al. Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis[J]. Cell, 2015, 160 (6): 1246- 1260.
doi: 10.1016/j.cell.2015.02.038 |
| 37 | 李晓娇, 何燕华, 朱新宇, 等. CRISPR/Cas9技术在猪、鸡中的应用研究进展[J]. 中国畜牧兽医, 2022, 49 (12): 4665- 4673. |
| LI X J , HE Y H , ZHU X Y , et al. Research progress on application of CRISPR/Cas9 technology in pigs and chickens[J]. China Animal Husbandry&, Veterinary Medicine, 2022, 49 (12): 4665- 4673. | |
| 38 |
GAO M , ZHU X , YANG G , et al. CRISPR/Cas9-mediated gene editing in porcine models for medical research[J]. DNA Cell Biol, 2021, 40 (12): 1462- 1475.
doi: 10.1089/dna.2020.6474 |
| 39 |
WANG H , SHEN L , CHEN J , et al. Deletion of CD163 exon 7 confers resistance to highly pathogenic porcine reproductive and respiratory viruses on pigs[J]. Int J Biol Sci, 2019, 15 (9): 1993- 2005.
doi: 10.7150/ijbs.34269 |
| 40 | BURKARD C , OPRIESSNIG T , MILEHAM A J , et al. Pigs lacking the scavenger receptor cysteine-rich domain 5 of CD163 are resistant to porcine reproductive and respiratory syndrome virus 1 infection[J]. J Virol, 2018, 92 (16): e00415- 18. |
| 41 |
KOSLOVÁ A , TREFIL P , MUCKSOVÁ J , et al. Precise CRISPR/Cas9 editing of the NHE1 gene renders chickens resistant to the J subgroup of avian leukosis virus[J]. Proc Natl Acad Sci U S A, 2020, 117 (4): 2108- 2112.
doi: 10.1073/pnas.1913827117 |
| 42 | 吴珊珊, 王学侨, 王鑫, 等. MSTN基因编辑鲁西牛屠宰性状与肉用品质分析[J]. 农业生物技术学报, 2023, 31 (1): 87- 97. |
| WU S S , WANG X Q , WANG X , et al. Analysis of slaughter traits and meat quality of MSTN gene-edited luxi cattle (bos taurus)[J]. Journal of Agricultural Biotechnology, 2023, 31 (1): 87- 97. | |
| 43 | WORKMAN A M , HEATON M P , VANDER LEY B L , et al. First gene-edited calf with reduced susceptibility to a major viral pathogen[J]. PNAS Nexus, 2023, 2 (5): 1- 14. |
| 44 | CHAN Y T , LU Y , WU J , et al. CRISPR-Cas9 library screening approach for anti-cancer drug discovery: overview and perspectives[J]. Theranostics, 2022, 12 (7): 3329- 3344. |
| 45 | 陈坤琳, 钱勇, 蒋临正, 等. SIRT7对牛乳腺上皮细胞乳蛋白、乳脂和乳糖合成关键基因表达的调控[J]. 南京农业大学学报, 2019, 42 (5): 917- 923. |
| CHEN K L , QIAN Y , JIANG L Z , et al. Effects of SIRT7 on the expression of key genes involved in lactoprotein, milk fat and lactose synthesis in dairy cow mammary epithelial cells[J]. Journal of Nanjing Agricultural University, 2019, 42 (5): 917- 923. |
| [1] | FU Yu, YANG Zhuo, ZHENG Hao, SUN Guohan, SHEN Wenjuan, HAN Xiaohong, TAO Jinzhong. Correlation between Related Factors in Peripheral Plasma and Pregnancy Status of Dairy Cows in Early Breeding Period [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(6): 2790-2800. |
| [2] | WANG Qinqian, GAO Zhendong, LU Ying, MA Ruoshan, DENG Weidong, HE Xiaoming. Research Progress of Whole Genome Resequencing in Chinese Indigenous Cattle [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(5): 2026-2037. |
| [3] | ZHANG Junxing, SHENG Hui, HAN Liyun, ZHANG Hailiang, ZHANG Yi, CAI Bei, MA Yun, WANG Yachun. The Impact of Health Events on Important Economic Traits in Holstein Lactating Cows [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(5): 2203-2218. |
| [4] | QIAO Yarui, MIAO Yuhang, HUANG Qian, ZHOU Xuezhang. Research on the Biological Characteristics of Enterococcus faecalis in Dairy Cow Mastitis in Ningxia [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(5): 2325-2339. |
| [5] | WANG Jinxiang, SU Jinbo, FU Huanru, SUN Shikun, GAO Chengfang, CHEN Dongjin, SANG Lei, XIE Xiping. Pathogenicity and Genomic Features of Rabbit Sourced Serogroup A Pasteurella multocida Isolates Pm3 and Pm6 [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(5): 2340-2352. |
| [6] | ZHAO Ying, WANG Jinglei, WANG Meng, WANG Libin, ZHANG Qian, LI Zhijie, MA Xin, YU Sijiu, PAN Yangyang. Preparation and Characterization of Forsythiaside A and Kaempferol Encapsulated in Milk-derived Exosomes and Evaluation of Anti-inflammatory Effects in vitro [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(5): 2481-2495. |
| [7] | MA Xiuling, ZHANG Xinru, CHEN Ying, LIANG Hongyan, ABDUREYIMU Gulimire, WANG Liqin, LIN Jiapeng, LI Weijian, WANG Xuguang, WU Yangsheng. PDGFD Gene Editing in Altay Sheep Embryos [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(4): 1700-1711. |
| [8] | FAN Manting, HUANG Ruoting, SHE Yuanhang, GUO Jianchao, LIU Jianying, GUO Yongqing. Research Progress on the Application of Omics Technology in the Pathogenesis and Diagnosis of Mastitis in Dairy Cows [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(3): 1076-1088. |
| [9] | WANG Zichen, ZHANG Na, ZHANG Wanting, ZHU Hao, LU Xubin, TIAN Yu, GE Jiwen, WANG Yongkuan, CHEN Yuhai, WANG Yachun, YANG Zhangping, MAO Yongjiang. Analysis of Influencing Factors and Estimation of Genetic Parameters on Daily Rumination Time and Daily Milk Yield in Holstein Cows [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(3): 1180-1188. |
| [10] | HU Xin, YOU Wei, JIANG Fugui, CHENG Haijian, SUN Zhigang, SONG Enliang. Analysis of Genetic Diversity and Population Structure of Simmental Cattle Based on Whole Genome Resequencing [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(3): 1189-1202. |
| [11] | ZHANG Shiqi, ZHENG Nan, WANG Jiaqi, ZHAO Shengguo. Effect of Dietary NFC/NDF Ratio on the Metabolic Flux of Microbial Urea Nitrogen in the Rumen of Dairy Cows [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(3): 1302-1312. |
| [12] | YANG Xiaowen, NING Wenqing, ZHOU Shizhong, YUAN Yaqin, HOU Xuexin, DING Jiabo. Establishment of a Quantitative Real-time PCR Detection Method for Trimethoprim-Sulfamethoxazole-resistance Strains of Brucella melitensis [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(3): 1465-1472. |
| [13] | XIE Yaru, JIN Haoyan, KONG Chen, CAI Bei, ZHANG Lingkai. Research Progress of CRISPR/Cas9 System in Livestock Germ Cells [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(2): 479-491. |
| [14] | WU Pingxian, WANG Junge, DIAO Shuqi, CHAI Jie, ZHA Lin, GUO Zongyi, CHEN Hongyue, LONG Xi. Analysis of Genetic Architecture Characteristics and Selection Signature by Imputed Whole Genome Sequencing Data in Rongchang Pigs [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(1): 147-158. |
| [15] | WANG Jing, GUAN Shuwen, ZHAO Xiaobo, WANG Linwei, GUO Gang, JIANG Linshu. The Protective Effect of Bamboo Leaf Flavonoids on H2O2-induced Pyroptosis in Bovine Mammary Epithelial Cells [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(1): 281-294. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||