





Acta Veterinaria et Zootechnica Sinica ›› 2025, Vol. 56 ›› Issue (6): 2639-2648.doi: 10.11843/j.issn.0366-6964.2025.06.010
• Animal Genetics and Breeding • Previous Articles Next Articles
WU Tong1( ), WANG Nan1, XING Yuxin1, ZHANG Ben1,2, HU Panyang1,2, ZHANG Haitao3, ZHU Yufeng1,2, WU Xiangzhe1,2, YANG Feng1,2, LI Xiuling1,2, WANG Kejun1,2, HAN Xuelei1,2, LI Xinjian4, YU Tong1,2, BAI Jun1,2, LI Gaiying1,2,*(
), WANG Nan1, XING Yuxin1, ZHANG Ben1,2, HU Panyang1,2, ZHANG Haitao3, ZHU Yufeng1,2, WU Xiangzhe1,2, YANG Feng1,2, LI Xiuling1,2, WANG Kejun1,2, HAN Xuelei1,2, LI Xinjian4, YU Tong1,2, BAI Jun1,2, LI Gaiying1,2,*( ), QIAO Ruimin1,2,*(
), QIAO Ruimin1,2,*( )
)
Received:2024-10-30
Online:2025-06-23
Published:2025-06-25
Contact:
LI Gaiying, QIAO Ruimin
E-mail:925998286@qq.com;ligaiying@126.com;ruiminqiao@henau.edu.cn
CLC Number:
WU Tong, WANG Nan, XING Yuxin, ZHANG Ben, HU Panyang, ZHANG Haitao, ZHU Yufeng, WU Xiangzhe, YANG Feng, LI Xiuling, WANG Kejun, HAN Xuelei, LI Xinjian, YU Tong, BAI Jun, LI Gaiying, QIAO Ruimin. Association Analysis of Backfat Thickness and Genome-Wide Copy Number Variations in Yunan Black Pigs[J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(6): 2639-2648.
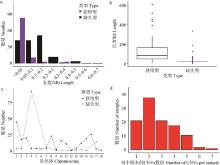
Fig. 1
Distribution of genome-wide copy number variation of Yunan black pigs a. The quantity distribution of CNV in different length intervals; b. Length characteristics of different types of CNV; c. The number and distribution of CNV on chromosomes; d. The quantity distribution of CNV in the sample"
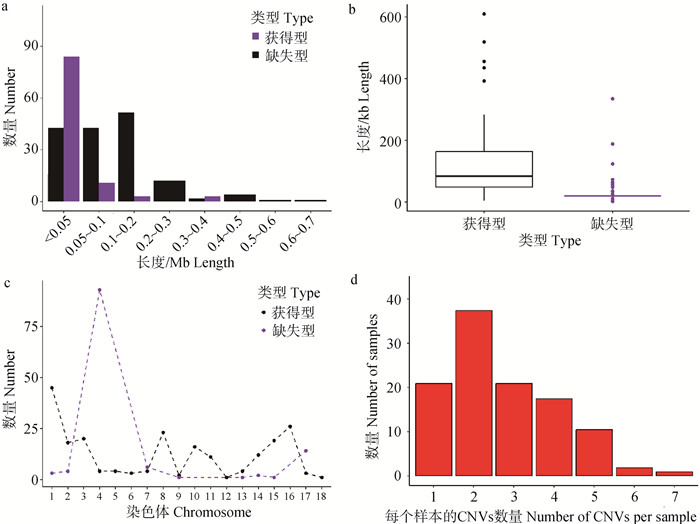
Table 1
Distribution of copy number variation regions on the autosomes of Yunan black pigs"
| 染色体 Chromosome | 拷贝数变异区域 Copy number variation region | |||||
| CNVR数目 CNVR count | 长度/bp Length | 染色体占比/% Percentage of chromosomes | 均值/bp Average size | 最大值/bp Max size | 最小值/bp Min size | |
| 1 | 6 | 419 684 | 0.153 | 69 947.333 | 188 012 | 12 589 |
| 2 | 5 | 882 284 | 0.581 | 176 456.800 | 609 138 | 11 884 |
| 3 | 8 | 1 115 033 | 0.839 | 139 379.125 | 518 657 | 14 568 |
| 4 | 1 | 19 611 | 0.015 | 19 611.000 | 19 611 | 19 611 |
| 5 | 3 | 524 569 | 0.502 | 174 856.333 | 434 828 | 31 410 |
| 6 | 2 | 179 723 | 0.105 | 89 861.500 | 95 517 | 84 206 |
| 7 | 4 | 377 690 | 0.310 | 94 422.500 | 187 907 | 38 779 |
| 8 | 6 | 891 973 | 0.642 | 148 662.167 | 455 579 | 27 728 |
| 9 | 3 | 265 452 | 0.190 | 88 484.000 | 189 777 | 25 963 |
| 10 | 1 | 83 312 | 0.120 | 83 312.000 | 83 312 | 83 312 |
| 11 | 5 | 455 969 | 0.576 | 91 193.800 | 271 456 | 16 937 |
| 12 | 1 | 7 074 | 0.011 | 7 074.000 | 7 074 | 7 074 |
| 13 | 3 | 282 199 | 0.135 | 94 066.333 | 183 842 | 36 129 |
| 14 | 3 | 158 815 | 0.112 | 52 938.333 | 62 587 | 48 045 |
| 15 | 6 | 457 949 | 0.326 | 76 324.833 | 116 540 | 34 047 |
| 16 | 2 | 164 561 | 0.206 | 82 280.500 | 146 652 | 17 909 |
| 17 | 2 | 441 155 | 0.695 | 220 577.500 | 342 967 | 98 188 |
| 18 | 1 | 3 918 | 0.007 | 3 918.000 | 3 918 | 3 918 |
Table 2
Descriptive statistics of backfat thickness phenotype of Yunan black pig population"
| 背膘厚表型 Average backfat thickness phenotype | 平均值/mm Average size | 标准差/mm S.D. | 最大值/mm Max size | 最小值/mm Min size | 变异系数/% C.V |
| P2点 P2 point | 36.058 | 7.983 | 53.900 | 20.700 | 22.139 |
| 肩部最厚处 Maximum shoulder thickness | 17.259 | 3.166 | 26.900 | 9.400 | 18.322 |
| 六七肋骨处 6th-7th rib interface | 38.243 | 9.313 | 60.300 | 20.700 | 24.351 |
| 最后肋骨处 The last rib | 33.218 | 7.693 | 53.900 | 18.200 | 23.159 |
| 腰荐结合处 Lumbosacral junction | 21.295 | 3.506 | 36.400 | 13.800 | 16.463 |
| 平均背膘厚 Average backfat thickness | 27.504 | 4.900 | 40.425 | 18.000 | 17.817 |
Table 3
Results of association study between CNVR and backfat thickness of Yunan black pig population"
| 背膘厚表型 Average backfat thickness phenotype | 染色体 Chromosome | CNV位置/bp Position | 类型 Category | CNV长度/bp Length | P值 P-value | 候选基因 Candidate gene |
| 腰荐结合处 Lumbosacral junction | 1 | 152 201 203~152 222 760 | Gain | 21 558 | 2.7×10-3 | SOCS6, PTTN, CD226 |
| P2点 P2 point | 8 | 79 697 483~79 725 210 | Loss | 27 728 | 1.52×10-3 | NR3C2, ARHGAP10, DCLK2, IQCM |
| 肩部最厚处 Maximum shoulder thickness | 8 | 79 697 483~79 725 210 | Loss | 27 728 | 6.4×10-3 | NR3C2, ARHGAP10, DCLK2, IQCM |
| 最后肋骨处 The last rib | 16 | 13 214 860~13 232 768 | Loss | 17 909 | 1.76×10-3 | CDH9, CDH10, U2, U6 |
| 1 | 李言, 黎观红, 周华, 等. 微生物发酵饲料在生猪生产中的应用[J]. 中国饲料, 2024 (15): 111- 117. |
| LI Y , LI G H , ZHOU H , et al. Application of microbial fermented feed in pig production[J]. China Feed, 2024 (15): 111- 117. | |
| 2 | 竹学军, 李孝法, 赵传发. 豫南黑猪培育历程及品种优势[J]. 中国牧业通讯, 2009 (4): 36- 38. |
| ZHU X J , LI X F , ZHAO C F . The breeding process and variety advantages of Yunnan Black Pig[J]. China Animal Husbandry and Veterinary Medicine Communication, 2009 (4): 36- 38. | |
| 3 | 张斌, 侯以强, 李平, 等. 豫南黑猪新品种培育经验[J]. 河南畜牧兽医(综合版), 2008 (11): 14- 15. |
| ZHANG B , HOU Y Q , LI P , et al. Experience in breeding the new variety of Yunnan Black Pig[J]. Henan Animal Husbandry and Veterinary Medicine (Comprehensive Edition), 2008 (11): 14- 15. | |
| 4 | LAVERY A , LAWLOR P G , MAGOWAN E , et al. An association analysis of sow parity, live-weight and back-fat depth as indicators of sow productivity[J]. Animal, 2018, 13 (3): 622- 630. |
| 5 |
WOONG K G . Analysis of carcass quality grades according to gender, backfat thickness and carcass weight in pigs[J]. J Anim Sci Technol, 2012, 54 (1): 29- 33.
doi: 10.5187/JAST.2012.54.1.29 |
| 6 |
VANBA H , HYUNWOO S , PILNAM S , et al. Back-fat thickness as a primary index reflecting the yield and overall acceptance of pork meat[J]. Anim Sci J, 2021, 92 (1): e13515.
doi: 10.1111/asj.13515 |
| 7 |
AHOUDE A , MÉTHOT S , MURPHY B D , et al. Relationships between backfat thickness and reproductive efficiency of sows: A two-year trial involving two commercial herds fixing backfat thickness at breeding[J]. Can J Anim Sci, 2010, 90 (3): 429- 436.
doi: 10.4141/CJAS09115 |
| 8 | FEUK L , CARSON A R , SCHERER S W . Structural variation in the human genome[J]. Nat Rev Genet, 2006, 7 (2): 85- 97. |
| 9 |
何阳花, 俞英, 张沅. 拷贝数变异与疾病的关系及其在动物抗病育种中的应用前景[J]. 遗传, 2008, 30 (11): 1385- 1391.
doi: 10.3321/j.issn:0253-9772.2008.11.002 |
|
HE Y H , YU Y , ZHANG Y . Relationship between copy number variation and disease and its application prospects in animal disease-resistant breeding[J]. Heredity, 2008, 30 (11): 1385- 1391.
doi: 10.3321/j.issn:0253-9772.2008.11.002 |
|
| 10 | 李大伟, 任稳稳, 喇永富, 等. 阿什旦牦牛MEF2A基因拷贝数变异与生长性状关联分析[J]. 中国草食动物科学, 2024, 44 (5): 84- 88. |
| LI D W , REN W W , LA Y F , et al. Association analysis of copy number variation of the MEF2A gene with growth traits in Ashidan yak[J]. Chinese Journal of Ruminant Science, 2024, 44 (5): 84- 88. | |
| 11 | 马丽娜, 刘永进, 马青, 等. 滩羊C1H3orf33和CNV14基因拷贝数变异与生长性状的关联研究[J]. 中国畜牧杂志, 2023, 59 (3): 123- 130. |
| MA L N , LIU Y J , MA Q , et al. Association study of copy number variation of the C1H3orf33 and CNV14 genes with growth traits in Tan sheep[J]. China Animal Husbandry Magazine, 2023, 59 (3): 123- 130. | |
| 12 | 吕世杰, 施巧婷, 冯亚杰, 等. SMG9基因拷贝数变异和不同因素对奶牛繁殖性状的影响[J]. 畜牧与兽医, 2022, 54 (5): 9- 13. |
| LÜ S J , SHI Q T , FENG Y J , et al. Effects of SMG9 gene copy number variation and different factors on reproductive traits in dairy cows[J]. Animal Husbandry and Veterinary Medicine, 2022, 54 (5): 9- 13. | |
| 13 |
祝雪丽, 张龙超, 王立贤, 等. 北京黑猪AQP9和RPS10基因多态性及其与背膘厚的关联分析[J]. 畜牧兽医学报, 2024, 55 (1): 87- 98.
doi: 10.11843/j.issn.0366-6964.2024.01.010 |
|
ZHU X L , ZHANG L C , WANG L X , et al. Polymorphism of AQP9 and RPS10 genes and their association with backfat thickness in Beijing Black pigs[J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55 (1): 87- 98.
doi: 10.11843/j.issn.0366-6964.2024.01.010 |
|
| 14 |
吴嘉浩, 吴姿仪, 窦腾飞, 等. 豫农黑猪生长相关性状的拷贝数变异全基因组关联分析研究[J]. 畜牧兽医学报, 2025, 56 (3): 1110- 1119.
doi: 10.11843/j.issn.0366-6964.2025.03.013 |
|
WU J H , WU Z Y , DOU T F , et al. Genome wide association analysis of copy number variation of growth related traits in Yunong black pig[J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56 (3): 1110- 1119.
doi: 10.11843/j.issn.0366-6964.2025.03.013 |
|
| 15 | CENDRON F , CASSANDRO M , PENASA M . Genome-wide investigation to assess copy number variants in the ltalian local chicken population[J]. JAnim Sci Biotechnol, 2024, 15 (1): 2. |
| 16 | DE OLIVEIRA H R , CHUD T C S , OLIVEIRA G AJR , et al. Genome-wide association analyses reveal copy number variant regions associated with reproduction and disease traits in Canadian Holstein cattle[J]. J Dairy Sci, 2024, 107 (9): 7052- 7063. |
| 17 | 邱恒清. 巴马香猪全基因组拷贝数变异的检测及其与五块四肢骨骼长度的关联分析[D]. 南昌: 江西农业大学, 2017. |
| QIU H Q. Detection of genome-wide copy number variation in Bama Xiang pigs and its association with the length of five limb bones[D]. Nanchang: Jiangxi Agricultural University, 2017. (in Chinese) | |
| 18 | 谢鑫峰, 王子轶, 钟梓奇, 等. 全基因组关联分析的扩展方法及其在畜禽中应用的研究进展[J]. 中国畜牧兽医, 2024, 51 (4): 1382- 1389. |
| XIE X F , WANG Z Y , ZHONG Z Q , et al. Advances in the extended methods of genome-wide association studies and their applications in livestock and poultry[J]. China Animal Husbandry and Veterinary Medicine, 2024, 51 (4): 1382- 1389. | |
| 19 | KAI W , MINGYAO L , DEXTER H , et al. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data[J]. Genome Res, 2007, 17 (11): 1665- 1674. |
| 20 | 王源. 猪繁殖性状全基因组关联分析及拷贝数变异检测[D]. 北京: 中国农业大学, 2018. |
| WANG Y. Genome-wide association analysis and copy number variation detection of reproductive traits in pigs[D]. Beijing: China Agricultural University, 2018. (in Chinese) | |
| 21 | 刘佳森, 张莉, 李蕴华, 等. 苏尼特羊拷贝数变异的基因组分布特征研究[J]. 中国畜牧兽医, 2013, 40 (10): 173- 178. |
| LIU J S , ZHANG L , LI Y H , et al. Genomic distribution characteristics of copy number variation in Sunite sheep[J]. China Animal Husbandry and Veterinary Medicine, 2013, 40 (10): 173- 178. | |
| 22 | VYKOUKALOVÁ Z , KNOLL A , AČG EPICA S . Porcine perilipin (PLIN) gene: Structure, polymorphism and association study in Large White pigs[J]. Czech J Anim Sci, 2018, 54 (8): 359- 364. |
| 23 | JIAO S , MALTECCA C , GRAY K A , et al. Feed intake, average daily gain, feed efficiency, and real-time ultrasound traits in Duroc pigs: Ⅱ. Genomewide association[J]. J Anim Sci, 2014, 92 (7): 2846- 2860. |
| 24 | LAKHSSASSI K , LAHOZ B , SARTO P , et al. Genome-wide association study demonstrates the role played by the CD226 gene in Rasa Aragonesa sheep reproductive seasonality[J]. Animals (Basel), 2021, 11 (4): 1171. |
| 25 | FOWLER K E , PONG-WONG R , BAUER J , et al. Genome-wide analysis reveals single nucleotide polymorphisms associated with fatness and putative novel copy number variants in three pig breeds[J]. BMC Genomics, 2013, 14, 784. |
| 26 | YIN C , WANG Y , ZHOU P , et al. Genomic scan for runs of homozygosity and selective signature analysis to identify candidate genes in Large White pigs[J]. Int J Mol Sci, 2023, 24 (16): 12914. |
| 27 | BOITARD S , LIAUBET L , PARIS C , et al. Whole-genome sequencing of cryopreserved resources from French Large White pigs at two distinct sampling times reveals strong signatures of convergent and divergent selection between the dam and sire lines[J]. Genet Sel Evol, 2023, 55 (1): 13. |
| 28 | RAMOS Z , GARRICK D J , BLAIR H T , et al. Genomic regions associated with wool, growth and reproduction traits in Uruguayan Merino sheep[J]. Genes (Basel), 2023, 14 (1): 167. |
| 29 | 张尾翼. 澳洲波尔山羊CNV鉴定及肌肉发育机制研究[D]. 重庆: 西南大学, 2023. |
| ZHANG W Y. CNV identification and muscle development mechanism of Boer goat in Australia[D]. Chongqing: Southwest University, 2023. (in Chinese) | |
| 30 |
邱恒清, 肖石军, 郭源梅. 利用猪1.4M高密度SNP芯片检测巴马香猪全基因组拷贝数变异[J]. 畜牧兽医学报, 2020, 51 (9): 2079- 2088.
doi: 10.11843/j.issn.0366-6964.2020.09.005 |
|
QIU H Q , XIAO S J , GUO Y M . Genome-wide copy number variation detection in Bama Xiang pigs using the 1.4M high-density SNP chip[J]. Acta Veterinaria et Zootechnica Sinica, 2020, 51 (9): 2079- 2088.
doi: 10.11843/j.issn.0366-6964.2020.09.005 |
|
| 31 | 路玉洁, 莫家远, 朱思燃, 等. 利用中芯一号SNP芯片检测隆林猪全基因组拷贝数变异[J]. 中国畜牧兽医, 2022, 49 (1): 23- 31. |
| LU Y J , MO J Y , ZHU S R , et al. Genome-wide copy number variation detection in Longlin pigs using the Zhongxin No.1 SNP chip[J]. China Animal Husbandry and Veterinary Medicine, 2022, 49 (1): 23- 31. | |
| 32 | FONTANESI L , BERETTI F , RIGGIO V , et al. Copy number variation and missense mutations of the Agouti signaling protein (ASIP) gene in goat breeds with different coat colors[J]. Cytogenet Genome Res, 2010, 126 (4): 333- 347. |
| 33 | 张力允, 黄智荣, 杨柳, 等. 二代基因组测序鉴别狮头鹅拷贝数变异及其与体重体尺关联[J]. 中国农业科学, 2024, 57 (14): 2892- 2903. |
| ZHANG L Y , HUANG Z R , YANG L , et al. Identification of copy number variation in Shitou geese by second-generation genome sequencing and its association with body weight and linear measurements[J]. Scientia Agricultura Sinica, 2024, 57 (14): 2892- 2903. | |
| 34 | 刘成铭, 杨祎挺, 甘麦邻, 等. 伍隍猪和内江猪拷贝数变异差异与嘴型全基因组关联分析[J]. 四川农业大学学报, 2023, 41 (6): 1090-1097, 147. |
| LIU C M , YANG Y T , GAN M L , et al. Differences in copy number variation between Wuhuang pigs and Neijiang pigs and genome-wide association analysis of snout shape[J]. Journal of Sichuan Agricultural University, 2023, 41 (6): 1090-1097+147. | |
| 35 | QIAN R , XIE F , ZHANG W , et al. Genome-wide detection of CNV regions between Anqing six-end-white and Duroc pigs[J]. Mol Cytogenet, 2023, 16 (1): 12. |
| 36 | PANDA S , KUMAR A , GAUR G K , et al. Genome-wide copy number variations using Porcine 60K SNP Beadchip in Landrace pigs[J]. Anim Biotechnol, 2023, 34 (6): 1891- 1899. |
| 37 | ZHANG F , GU W , HURLES M E , et al. Copy number variation in human health, disease, and evolution[J]. Annu Rev Genomics Hum Genet, 2009, 10 (1): 451- 481. |
| [1] | HUANG Yani, TANG Xi, LI Jingquan, WEI Jiacheng, WU Zhenfang, LI Xinyun, XIAO Shijun, ZHANG Zhiyan. Large-scale Population Analysis of Potential Causal Genes for Daily Weight Gain and Age at 100 kg in Pigs [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(3): 1100-1109. |
| [2] | WU Jiahao, WU Ziyi, DOU Tengfei, BAI Liyao, ZHANG Yongqian, DONG Lianhe, LI Pengfei, LI Xinjian, HAN Xuelei, LI Xiuling. Genome-wide Association Study of Copy Number Variation in Growth-Related Traits of Yunong-Black Pigs [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(3): 1110-1119. |
| [3] | PENG Peiya, CHEN Yuhan, YANG Long, WANG Ming, ZHAO Ruiting, HE Jun, YIN Yulong, LIU Mei. Research Progress of Copy Number Variation in Livestock [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(4): 1356-1369. |
| [4] | CAO Yuzhu, XING Yuxin, MA Chenglin, GUAN Hongbo, JIA Qihui, KANG Xiangtao, TIAN Yadong, LI Zhuanjian, LIU Xiaojun, LI Hong. Biological Characterization of Chicken FGF6 Gene and Association of Its Polymorphisms with Economic Traits [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(4): 1536-1550. |
| [5] | XU Donghui, XU Yuhui, LI Ruizhe, CHENG Haijian, MA Zhijie. Research Progress of Genome Copy Number Variations in Yak [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(3): 933-943. |
| [6] | ZHONG Xin, ZHANG Hui, ZHANG Chong, LIU Xiaohong. Research Progress on Genetic Breeding of Reproductive Performance in Sows [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(2): 438-450. |
| [7] | QI Junying, PEI Quanbang, ZHANG Wenkui, XU Teng, ZUO Mingxing, HAN Buying, LI Xue, LIU Dehui, WANG Song, ZHOU Baicheng, ZHAO Kai, TIAN Dehong. Genome-wide Selective Signal Identification and Association Analysis of Candidate Genes for Tibetan Sheep Wool Traits [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(12): 5511-5526. |
| [8] | ZHU Xueli, ZHANG Longchao, WANG Lixian, PU Lei, LIU Xin. Association Analysis of AQP9 and RPS10 Gene Polymorphisms with Backfat Thickness in Beijing Black Pigs [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(1): 87-98. |
| [9] | TANG Xinxin, ZHENG Jumei, LUO Na, YING Fan, ZHU Dan, LI Sen, LIU Dawei, AN Bingxing, WEN Jie, ZHAO Guiping, LI Hegang. Genetic Mechanism of Broiler Leg Disease Based on Genome-Wide Association Analysis [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(1): 99-109. |
| [10] | LIN Yan, HUANG Min, LI Xiujin, ZHANG Xumeng, HUANG Yunmao, TIAN Yunbo, WU Zhongping. Uncovering Genome-wide Copy Number Variations in 8 Duck Breeds Using Whole Genome Resequencing Data [J]. Acta Veterinaria et Zootechnica Sinica, 2023, 54(9): 3700-3709. |
| [11] | WU Jun, CAI Xiaodian, LIN Qing, ZHONG Zhanming, YE Haoqiang, WEI Chen, XU Zhiting, WU Xibo, SI Jinglei, ZHANG Zhe, LI Jiaqi. Weighted Single-step GWAS of Eye Muscle Area, Predicted Lean Meat Percentage and Average Backfat Thickness in A Yorkshire Pig Population [J]. Acta Veterinaria et Zootechnica Sinica, 2023, 54(4): 1403-1414. |
| [12] | BI Yazhen, SHANG Mingyu, HU Wenping, ZHANG Li. Correlation and Regression Analysis among Growth Traits and Association Analysis of TRHDE Gene Polymorphism with Growth Traits in Sheep [J]. Acta Veterinaria et Zootechnica Sinica, 2023, 54(4): 1415-1428. |
| [13] | SU Yanfang, YANG Man, NIU Naiqi, HOU Xinhua, ZHANG Longchao. Association Analysis of FABP3 and SCD Gene Polymorphisms with Meat Quality Traits in Beijing Black Pigs [J]. Acta Veterinaria et Zootechnica Sinica, 2023, 54(3): 966-975. |
| [14] | ZHANG Zhengkai, LI Yefang, YE Shaohui, JIANG Lin, MA Yuehui. Research Progress of Environmental Adaptability in Goats [J]. Acta Veterinaria et Zootechnica Sinica, 2022, 53(7): 2035-2046. |
| [15] | TIAN Weilong, LAN Ganqiu, ZHANG Longchao, WANG Lixian, LIANG Jing, LIU Xin. Detection of DKK3 and CCR1 Genes Polymorphisms and Their Association with Backfat Thickness in Beijing Black Pigs [J]. Acta Veterinaria et Zootechnica Sinica, 2022, 53(7): 2083-2093. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||