





Acta Veterinaria et Zootechnica Sinica ›› 2025, Vol. 56 ›› Issue (3): 1100-1109.doi: 10.11843/j.issn.0366-6964.2025.03.012
• Animal Genetics and Breeding • Previous Articles Next Articles
HUANG Yani1( ), TANG Xi1, LI Jingquan1, WEI Jiacheng1, WU Zhenfang2, LI Xinyun3, XIAO Shijun1,*(
), TANG Xi1, LI Jingquan1, WEI Jiacheng1, WU Zhenfang2, LI Xinyun3, XIAO Shijun1,*( ), ZHANG Zhiyan1,*(
), ZHANG Zhiyan1,*( )
)
Received:2024-07-26
Online:2025-03-23
Published:2025-04-02
Contact:
XIAO Shijun, ZHANG Zhiyan
E-mail:yani720@163.com;shjx_jxau@hotmail.com;bioducklily@hotmail.com
CLC Number:
HUANG Yani, TANG Xi, LI Jingquan, WEI Jiacheng, WU Zhenfang, LI Xinyun, XIAO Shijun, ZHANG Zhiyan. Large-scale Population Analysis of Potential Causal Genes for Daily Weight Gain and Age at 100 kg in Pigs[J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(3): 1100-1109.

Fig. 1
The density plot of SNPs imputed using the 50K chip across the genome, along with the PCA analysis plot A. Density plot of SNP across the genome after imputed; B. The PCA plot displays the first two principal components, red points represent Yorkshire pigs, green points represent Duroc pigs, and blue points represent Landrace pigs"

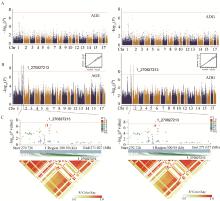
Fig. 2
Manhattan plots of GWAS before and after imputation for traits AGE and ADG, along with LD heatmap of significant loci A. Manhattan plots of GWAS analysis before imputation for AGE (left) and ADG (right), with significant threshold indicated by red line (P=5×10-8); B. Manhattan plots and Q-Q plot of GWAS analysis after imputation for AGE and ADG, with significant threshold indicated by red line (P=5×10-8); C. Local Manhattan plot (top) and linkage disequilibrium (LD) heatmap (bottom) of a critical region spanning 270627213 to 271027213 on chromosome 1, where increasing red color indicates higher linkage between SNP pairs"


Fig. 3
Genotype and candidate genes at significant locus 1_270827213 A. LocusZooms-like plot for the 200 kb upstream and downstream region of the significant locus 1_270827213; B. Phenotypic differences in AGE (top) and ADG (bottom) traits among different genotypes of the 1_270827213 locus in 4 501 individuals, using Kruskal-Wallis test, where P < 0.05 indicates significant genotype-phenotype differences; C. GO enrichment analysis of genes within a 1 Mb region flanking the significant locus 1_270827213"
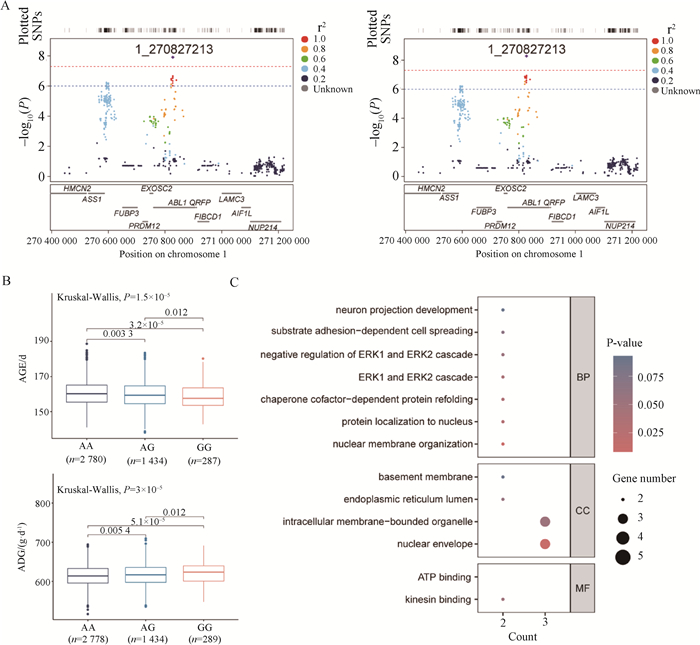
Table 4
GWAS and eQTL colocalization for the AGE trait, with positive (PP.H4>0.75) co-localization results"
| 组织 Tissue | 基因 Gene | SNP数量 SNP number | PP.H0.abf | PP.H1.abf | PP.H2.abf | PP.H3.abf | PP.H4.abf |
| 血液Blood | SPTAN1 | 49 | 0.036 909 816 | 0.010 126 732 | 0.101 001 093 | 0.026 886 008 | 0.825 076 351 |
| 血液Blood | DYNC212 | 27 | 0.097 667 167 | 0.017 861 012 | 0.108 656 142 | 0.019 113 934 | 0.756 701 746 |
| 大脑Brain | GLE1 | 69 | 5.87×10-5 | 1.43×10-5 | 0.058 270 568 | 0.013 224 164 | 0.928 432 334 |
| 大脑Brain | CRAT | 75 | 0.002 547 613 | 0.001 575 547 | 0.059 611 937 | 0.035 966 132 | 0.900 298 771 |
| 大脑Brain | TORA1 | 48 | 0.000 407 217 | 0.000 101 622 | 0.006 065 363 | 0.000 520 716 | 0.992 905 082 |
| 大脑Brain | GPR107 | 15 | 0.153 769 887 | 0.034 532 902 | 0.033 007 367 | 0.006 640 586 | 0.772 049 258 |
| 肝Liver | EXOSC2 | 55 | 0.068 160 145 | 0.051 873 945 | 0.050 815 144 | 0.037 882 094 | 0.791 268 673 |
| 肌肉Muscle | ENDOG | 25 | 0.000 221 142 | 0.000 108 116 | 0.016 117 346 | 0.006 903 087 | 0.976 650 309 |
| 肌肉Muscle | NUP188 | 3 | 0.090 167 323 | 0.000 845 334 | 0.143 226 173 | 0.000 577 585 | 0.765 183 585 |
| 脾Spleen | USP20 | 142 | 7.18×10-5 | 0.006 173 712 | 0.001 301 539 | 0.111 076 123 | 0.881 376 855 |
| 睾丸Testis | DYNC212 | 82 | 0.000 440 394 | 0.000 372 687 | 0.102 408 901 | 0.085 853 531 | 0.810 924 486 |
Table 5
GWAS and eQTL colocalization for the ADG trait, with positive (PP.H4>0.75) co-localization results"
| 组织 Tissue | 基因 Gene | SNP数量 SNP number | PP.H0.abf | PP.H1.abf | PP.H2.abf | PP.H3.abf | PP.H4.abf |
| 大脑Brain | CRAT | 75 | 0.007 236 723 | 0.002 530 961 | 0.169 333 086 | 0.058 459 875 | 0.762 439 354 |
| 大脑Brain | TOR1A | 48 | 0.000 187 523 | 0.000 111 277 | 0.002 793 096 | 0.000 661 182 | 0.996 246 923 |
| 大脑Brain | GPR107 | 15 | 0.068 069 088 | 0.037 270 208 | 0.014 611 322 | 0.007 127 288 | 0.872 922 094 |
| 肝Liver | EXOSC2 | 55 | 0.054 799 403 | 0.074 246 459 | 0.040 854 366 | 0.054 577 125 | 0.775 522 646 |
| 肌肉Muscle | TMEFF1 | 393 | 1.24×10-12 | 1.43×10-12 | 0.105 640 671 | 0.120 898 331 | 0.773 460 997 |
| 肌肉Muscle | ENDOG | 25 | 0.000 349 317 | 0.000 102 806 | 0.025 459 119 | 0.006 525 219 | 0.967 563 539 |
| 肌肉Muscle | PTGES | 82 | 4.09×10-7 | 7.29×10-8 | 0.161 141 799 | 0.027 926 643 | 0.810 931 076 |
| 肌肉Muscle | HMCN2 | 23 | 0.042 582 626 | 0.021 305 938 | 0.080 703 68 | 0.039 563 714 | 0.815 844 042 |
| 肌肉Muscle | ASS1 | 1 | 0.061 347 848 | 0.040 059 076 | 0.001 374 032 | 0 | 0.897 219 043 |
| 小肠 Small intestine | FNBP1 | 8 | 0.151 455 241 | 0.054 482 732 | 0.019 121 876 | 0.006 109 849 | 0.768 830 301 |
| 脾Spleen | USP20 | 142 | 0.000 256 969 | 0.006 037 688 | 0.004 660 025 | 0.108 610 336 | 0.880 434 982 |
| 1 | 张海峰, 王祖力, 陈泽芳, 等. 2023年全球生猪产业发展情况及2024年的趋势[J]. 猪业科学, 2024, 41 (2): 32- 36. |
| ZHANG H F , WANG Z L , CHEN Z F , et al. The global pig industry development in 2023 and trends for 2024[J]. Swine Industry Science, 2024, 41 (2): 32- 36. | |
| 2 |
JIANG Y , TANG S , WANG C , et al. A genome-wide association study of growth and fatness traits in two pig populations with different genetic backgrounds[J]. J Anim Sci, 2018, 96 (3): 806- 816.
doi: 10.1093/jas/skx038 |
| 3 |
TANG Z S , XU J Y , YIN L L , et al. Genome-wide association study reveals candidate genes for growth relevant traits in pigs[J]. Front Genet, 2019, 10, 302.
doi: 10.3389/fgene.2019.00302 |
| 4 |
HERRERA-CÁCERES W , SÁNCHEZ J P . Selection for feed efficiency using the social effects animal model in growing Duroc pigs: evaluation by simulation[J]. Genet Sel Evol, 2020, 52 (1): 53.
doi: 10.1186/s12711-020-00572-4 |
| 5 |
RUAN D L , ZHUANG Z W , DING R R , et al. Weighted single-step GWAS identified candidate genes associated with growth traits in a duroc pig population[J]. Genes (Basel), 2021, 12 (1): 117.
doi: 10.3390/genes12010117 |
| 6 | 许迪, 颜港, 张帅, 等. 大白猪生长性状影响因素分析及遗传参数估计[J]. 中国畜牧兽医, 2024, 51 (1): 193- 202. |
| XU D , YAN G , ZHANG S , et al. Influencing factors analysis and genetic parameters estimation of growth traits in yorkshire pigs[J]. China Animal Husbandry & Veterinary Medicine, 2024, 51 (1): 193- 202. | |
| 7 |
张笑科, 廖伟莉, 陈信佑, 等. 杜洛克猪生长性状全基因组关联分析及候选基因鉴定[J]. 畜牧兽医学报, 2023, 54 (5): 1868- 1876.
doi: 10.11843/j.issn.0366-6964.2023.05.010 |
|
ZHANG X K , LIAO W L , CHEN X Y , et al. Genome-wide association study for identifying candidate genes of growth traits in Duroc pigs[J]. Acta Veterinaria et Zootechnica Sinica, 2023, 54 (5): 1868- 1876.
doi: 10.11843/j.issn.0366-6964.2023.05.010 |
|
| 8 |
贺婕妤, 王斌虎, 廖柱, 等. 长白和大白猪主要生长性状的遗传参数估计[J]. 畜牧兽医学报, 2021, 52 (8): 2115- 2123.
doi: 10.11843/j.issn.0366-6964.2021.08.005 |
|
HE J Y , WANG B H , LIAO Z , et al. Estimation of genetic parameters of main growth traits in landrace and large white pigs[J]. Acta Veterinaria et Zootechnica Sinica, 2021, 52 (8): 2115- 2123.
doi: 10.11843/j.issn.0366-6964.2021.08.005 |
|
| 9 |
LOOS R J F , YEO G S H . The genetics of obesity: from discovery to biology[J]. Nat Rev Genet, 2022, 23 (2): 120- 133.
doi: 10.1038/s41576-021-00414-z |
| 10 |
NICA A C , MONTGOMERY S B , DIMAS A S , et al. Candidate causal regulatory effects by integration of expression QTLs with complex trait genetic associations[J]. PLoS Genet, 2010, 6 (4): e1000895.
doi: 10.1371/journal.pgen.1000895 |
| 11 |
PURCELL S , NEALE B , TODD-BROWN K , et al. PLINK: a tool set for whole-genome association and population-based linkage analyses[J]. Am J Hum Genet, 2007, 81 (3): 559- 575.
doi: 10.1086/519795 |
| 12 |
HOFMEISTER R J , RIBEIRO D M , RUBINACCI S , et al. Accurate rare variant phasing of whole-genome and whole-exome sequencing data in the UK Biobank[J]. Nat Genet, 2023, 55 (7): 1243- 1249.
doi: 10.1038/s41588-023-01415-w |
| 13 |
BROWNING B L , ZHOU Y , BROWNING S R . A one-penny imputed genome from next-generation reference panels[J]. Am J Hum Genet, 2018, 103 (3): 338- 348.
doi: 10.1016/j.ajhg.2018.07.015 |
| 14 |
TONG X K , CHEN D , HU J C , et al. Accurate haplotype construction and detection of selection signatures enabled by high quality pig genome sequences[J]. Nat Commun, 2023, 14 (1): 5126.
doi: 10.1038/s41467-023-40434-3 |
| 15 |
ZHOU X , STEPHENS M . Genome-wide efficient mixed-model analysis for association studies[J]. Nat Genet, 2012, 44 (7): 821- 824.
doi: 10.1038/ng.2310 |
| 16 |
JANNOT A S , EHRET G , PERNEGER T . P < 5×10-8 has emerged as a standard of statistical significance for genome-wide association studies[J]. J Clin Epidemiol, 2015, 68 (4): 460- 465.
doi: 10.1016/j.jclinepi.2015.01.001 |
| 17 |
DONG S S , HE W M , JI J J , et al. LDBlockShow: a fast and convenient tool for visualizing linkage disequilibrium and haplotype blocks based on variant call format files[J]. Brief Bioinform, 2021, 22 (4): bbaa227.
doi: 10.1093/bib/bbaa227 |
| 18 |
QUINLAN A R , HALL I M . BEDTools: a flexible suite of utilities for comparing genomic features[J]. Bioinformatics, 2010, 26 (6): 841- 842.
doi: 10.1093/bioinformatics/btq033 |
| 19 |
SHERMAN B T , HAO M , QIU J , et al. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update)[J]. Nucleic Acids Res, 2022, 50 (W1): W216- W221.
doi: 10.1093/nar/gkac194 |
| 20 |
GIAMBARTOLOMEI C , VUKCEVIC D , SCHADT E E , et al. Bayesian Test for colocalisation between Pairs of genetic association studies using summary statistics[J]. PLoS Genet, 2014, 10 (5): e1004383.
doi: 10.1371/journal.pgen.1004383 |
| 21 |
TENG J Y , GAO Y H , YIN H W , et al. A compendium of genetic regulatory effects across pig tissues[J]. Nat Genet, 2024, 56 (1): 112- 123.
doi: 10.1038/s41588-023-01585-7 |
| 22 |
GUI J X , YANG X Y , TAN C , et al. A cross-tissue transcriptome-wide association study reveals novel susceptibility genes for migraine[J]. J Headache Pain, 2024, 25 (1): 94.
doi: 10.1186/s10194-024-01802-6 |
| 23 | 邹若男, 时坤鹏, 张志勇, 等. 杜洛克猪生长性状全基因组关联分析[J/OL]. 中国畜牧杂志, (2024-04-18). https://doi.org/10.19556/j.0258-7033.20230629-08. |
| ZOU R N, SHI K P, ZHANG Z Y, et al. Genome-wide association analysis of growth traits in Duroc pigs[J/OL]. Chinese Journal of Animal Science, (2024-04-18). https://doi.org/10.19556/j.0258-7033.20230629-08. (in Chinese) | |
| 24 | 窦腾飞, 吴姿仪, 白利瑶, 等. 全基因组关联分析鉴定大白猪生长性状遗传变异及候选基因[J]. 中国畜牧杂志, 2023, 59 (8): 264- 272. |
| DOU T F , WU Z Y , BAI L Y , et al. Genome-wide association analysis identifies genetic variations and candidate genes for growth traits in Large White pigs[J]. Chinese Journal of Animal Science, 2023, 59 (8): 264- 272. | |
| 25 |
ZHOU S P , DING R R , MENG F M , et al. A meta-analysis of genome-wide association studies for average daily gain and lean meat percentage in two Duroc pig populations[J]. BMC Genomics, 2021, 22 (1): 12.
doi: 10.1186/s12864-020-07288-1 |
| 26 |
JUNG J H , LEE S M , OH S H . A genome-wide association study on growth traits of Korean commercial pig breeds using Bayesian methods[J]. Anim Biosci, 2024, 37 (5): 807- 816.
doi: 10.5713/ab.23.0443 |
| 27 | 王文斌. 基于低深度重测序挖掘猪经济性状相关突变位点[D]. 武汉: 华中农业大学, 2023. |
| WANG W B. Mining of mutation sites related to economic traits of pigs based on low depth resequencing[D]. Wuhan: Huazhong Agricultural University, 2023. (in Chinese) | |
| 28 |
VIOLANTE S , IJLST L , RUITER J , et al. Substrate specificity of human carnitine acetyltransferase: implications for fatty acid and branched-chain amino acid metabolism[J]. Biochim Biophys Acta, 2013, 1832 (6): 773- 779.
doi: 10.1016/j.bbadis.2013.02.012 |
| 29 |
MUOIO D M , NOLAND R C , KOVALIK J P , et al. Muscle-specific deletion of carnitine acetyltransferase compromises glucose tolerance and metabolic flexibility[J]. Cell Metab, 2012, 15 (5): 764- 777.
doi: 10.1016/j.cmet.2012.04.005 |
| 30 | FEITOSA N M , ZHANG J L , CARNEY T J , et al. Hemicentin 2 and Fibulin 1 are required for epidermal-dermal junction formation and fin mesenchymal cell migration during zebrafish development[J]. Dev Biol, 2012, 369 (2): 235- 248. |
| 31 | SAMPEY A V , MONRAD S , CROFFORD L J . Microsomal prostaglandin E synthase-1:the inducible synthase for prostaglandin E2[J]. Arthritis Res Ther, 2005, 7 (3): 114- 117. |
| 32 |
LU X Y , SHI X J , HU A , et al. Feeding induces cholesterol biosynthesis via the mTORC1-USP20-HMGCR axis[J]. Nature, 2020, 588 (7838): 479- 484.
doi: 10.1038/s41586-020-2928-y |
| 33 |
TSUJITA K , SUETSUGU S , SASAKI N , et al. Coordination between the actin cytoskeleton and membrane deformation by a novel membrane tubulation domain of PCH proteins is involved in endocytosis[J]. J Cell Biol, 2006, 172 (2): 269- 279.
doi: 10.1083/jcb.200508091 |
| 34 |
YOSTEN G L C , REDLINGER L J , SAMSON W K . Evidence for an interaction of neuronostatin with the orphan G protein-coupled receptor, GPR107[J]. Am J Physiol Regul Integr Comp Physiol, 2012, 303 (9): R941- R949.
doi: 10.1152/ajpregu.00336.2012 |
| 35 |
SAMSON W K , ZHANG J V , AVSIAN-KRETCHMER O , et al. Neuronostatin encoded by the somatostatin gene regulates neuronal, cardiovascular, and metabolic functions[J]. J Biol Chem, 2008, 283 (46): 31949- 31959.
doi: 10.1074/jbc.M804784200 |
| [1] | WU Jiahao, WU Ziyi, DOU Tengfei, BAI Liyao, ZHANG Yongqian, DONG Lianhe, LI Pengfei, LI Xinjian, HAN Xuelei, LI Xiuling. Genome-wide Association Study of Copy Number Variation in Growth-Related Traits of Yunong-Black Pigs [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(3): 1110-1119. |
| [2] | YANG Yuting, CHEN Guoliang, CHANG Qiaoning, BAO Wu, LIU Jingchao, JI Mengting, RONG Xiaoyin, GUO Xiaohong, YANG Yang, LI Bugao. miR-375-3p Targets Fam229a to Regulate Porcine Precursor Adipocyte Differentiation [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(3): 1120-1133. |
| [3] | JIA Wanli, WANG Jiying, LI Jingxuan, WANG Yanping, GENG Liying, ZHANG Chuansheng, ZHAO Xueyan. Identification of Key Genes Affecting Drip Loss in Laiwu Pigs Based on Transcriptome Sequencing Technology [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(3): 1134-1146. |
| [4] | PAN Junyi, WU Qingyao, TAN Bi'e, GUO Qiuping, HUANG Ruilin, CHEN Jiashun. Research Progress on Precise Nutrition Supply Technology and Intelligent Farming Equipment for Growing-Finishing Pigs [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(2): 501-512. |
| [5] | XIE Jingfei, ZHANG Yibo, ZHAO Yang, LIU Kaili, HUANG Shucheng. Research Progress of Food Intolerance in Cats [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(1): 82-94. |
| [6] | ZHOU Xianshan, HUANG Shihui, NIU Xi, RAN Xueqin, WANG Jiafu. Differential Expression Study of Structural Variation in the Ubiquitin Ligase 2 Gene of Xiang Pigs with Wrinkled Skin [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(1): 136-146. |
| [7] | WU Shuang, YIN Na, YU Mohan, PING Yuyu, BAI Hao, CHEN Shihao, CHANG Guobin. The Effect of TRIM39.2 Overexpression on the Transcriptional Expression of Chicken Macrophages [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(1): 178-188. |
| [8] | LU Xiu, ZHANG Ming'ai, KONG Min, ZHANG Jing, WANG Binghan, HOU Zhongyi, TENG Xingyi, JIANG Yajing, FAN Wenlei, WANG Baowei. Screening for Candidate Genes Related to Egg Production in Wulong Geese Based on Transcriptome and Proteome Analyses [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(1): 232-245. |
| [9] | Dong CHEN, Wenxuan ZHOU, Zhenjian ZHAO, Qi SHEN, Yang YU, Shengdi CUI, Junge WANG, Ziyang CHEN, Shixin YU, Jiamiao CHEN, Xiangfeng WANG, Pingxian WU, Zongyi GUO, Jinyong WANG, Guoqing TANG. Development of a Pig Intramuscular Fat Content and Eye Muscle Area Measurement System Based on Computer Vision Technology [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(9): 3843-3852. |
| [10] | Wei LIU, Jiayi MA, Haoyu GENG, Tian XIE, Sunan MIAO, Zongjie LIAO, Shizhong GENG. Isolation, Identification and Characterization of a Broad Spectrum Salmonella Phage [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(9): 4061-4068. |
| [11] | Meila NA, Kenan LI, Haidong DU, Wenliang GUO, Renhua NA. Study on the Differences of Fungal Diversity in Rumen and Feces of Inner Mongolia Cashmere Goats at Different Ages [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(8): 3526-3540. |
| [12] | Wenjun XU, Nan ZHENG, Jiaqi WANG, Lu MENG. Study on the Community Structure of Psychrophilic Bacteria in Raw Milk from North Regions of China based on Metagenomic Sequencing [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(8): 3659-3668. |
| [13] | Huihui DUAN, Shihang REN, Hongyin ZHANG, Rui YU, Zhonghu LIU, Xiangdang DU, Yanhong SHANG. Horizontal Transfer of Drug Resistance Genes Carried by ICE_Prophage in Streptococcus suis SC124 [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(8): 3699-3705. |
| [14] | Ya’nan LI, Tianwen MA, Yuhui MA, Chengwei WEI. Bilobalide Regulates Mitochondrial Biogenesis Mediated by AMPK-SIRT3 Positive Feedback Loop and Improves Inflammatory Damage of ATDC5 Chondrocytes [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(8): 3714-3724. |
| [15] | Yingguang LÜ, Guangming JIAO, Jinfang SANG, Zhipeng KOU, Tao LIU, Yue WANG, Xiangyu LU, Chenxi PIAO, Yajun MA, Jiantao ZHANG, Hongbin WANG. The Effect of Adipose Mesenchymal Stem Cells on the Healing Process of Autologous Skin Transplantation in Bama Miniature Pigs [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(7): 3193-3204. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||