





Acta Veterinaria et Zootechnica Sinica ›› 2025, Vol. 56 ›› Issue (4): 1700-1711.doi: 10.11843/j.issn.0366-6964.2025.04.019
• Animal Genetics and Breeding • Previous Articles Next Articles
MA Xiuling1( ), ZHANG Xinru2, CHEN Ying2, LIANG Hongyan2, ABDUREYIMU Gulimire2, WANG Liqin2, LIN Jiapeng2, LI Weijian1, WANG Xuguang1,*(
), ZHANG Xinru2, CHEN Ying2, LIANG Hongyan2, ABDUREYIMU Gulimire2, WANG Liqin2, LIN Jiapeng2, LI Weijian1, WANG Xuguang1,*( ), WU Yangsheng2,*(
), WU Yangsheng2,*( )
)
Received:2024-08-28
Online:2025-04-23
Published:2025-04-28
Contact:
WANG Xuguang, WU Yangsheng
E-mail:2234594165@qq.com;wangxuguang@xjau.edu.cn;xj_wys@126.com
CLC Number:
MA Xiuling, ZHANG Xinru, CHEN Ying, LIANG Hongyan, ABDUREYIMU Gulimire, WANG Liqin, LIN Jiapeng, LI Weijian, WANG Xuguang, WU Yangsheng. PDGFD Gene Editing in Altay Sheep Embryos[J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(4): 1700-1711.
Table 1
sgRNA sequences at 3 loci of sheep PDGFD"
| 序号 Number | sgRNA名称 Name of sgRNA | 核心序列 Core sequence | 基因位置 Location of genes |
| 1 | PDGFD-314-sgRNA2 | GGACTAATATCAGTGAAACG | Oar_v3.1,Chr15:3859314 |
| 2 | PDGFD-314-sgRNA5 | CTAATATCAGTGAAACGGGG | Oar_v3.1,Chr15:3859314 |
| 3 | PDGFD-410-sgRNA | GACGTACCACTCTGAGCACA | Oar_v.4.0, Chr15:3904410 |
| 4 | PDGFD-397-sgRNA2 | TCAGTGCCTGGCTCAAATAG | Oar_Rambouillet v1.0, Chr15:4172397 |
| 5 | PDGFD-397-sgRNA7 | TGGCTCAAATAGCGGCTTGT | Oar_Rambouillet v1.0, Chr15:4172397 |
Table 2
Primers used for PCR amplification"
| 序号 Number | 引物名称 Primer name | 引物序列 Primer sequence | 产物/bp Product |
| 1 | PDGFD-410-F1 | TCTGTTGTCGCTTAGAGTTT | 616 |
| 2 | PDGFD-410-F2 | AAAAGTGAAAGGGAAGTCG | 415 |
| 3 | PDGFD-410-R | TAGAAGGATGGCAGAAAGG | |
| 4 | PDGFD-314-F1 | AAAATGAGTGGTATGTGGGTG | 597 |
| 5 | PDGFD-314-F2 | GGGCTAAGGTCCCTGGAGT | 409 |
| 6 | PDGFD-314-R | GGGAAATTCTTGTGGCAGT | |
| 7 | PDGFD-397-F | GATTGCCCTCCAGATACCA | |
| 8 | PDGFD-397-R1 | GAGTGGTCTAAGCGGTTTT | 746 |
| 9 | PDGFD-397-R2 | CTGCTTCATTCCACCTTCC | 474 |
| 10 | PDGFD-397-HIT-F | ggagtgagtacggtgtgcGATTGCCCTCCAGATACCA | 262 |
| 11 | PDGFD-397-HIT-R | gagttggatgctggatggGGACTGTCCCTCTTTGGGT |
Table 3
Base types of PDGFD gene SNP loci in Altay sheep genome"
| 位点 Site | PDGFD SNP位点[文献] SNP loci in PDGFD | 哈萨克羊对应位置及碱基类型 Kazakh sheep corresponding position and base type |
| 1 | Oar_v3.1:Chr15:3622093(C>T)[ | 3 817 380 (T) |
| 2 | Oar_v3.1:Chr15:3859314(T>C)[ | 4 066 876 (C) |
| 3 | Oar_v3.1:Chr15:4031794(A>C)[ | 4 242 267 (G) |
| 4 | Oar_v.4.0:Chr15:3852134(C>T)[ | 4 055 473 (C) |
| 5 | Oar_v.4.0:Chr15:3904410(G>A)[ | 4 107 625 (A) |
| 6 | Oar_v.4.0:Chr15:4122606(C>G)[ | 4 326 797 (G) |
| 7 | Oar_Rambouillet v1.0:Chr15:4172397(A>G)[ | 3 827 484 (G) |
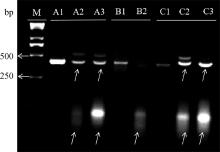
Fig. 1
Electrophoresis patterns of PCR amplified enzyme digestion products for sgRNA M.Trans2K Plus Ⅱ DNA Marker; A1. PDGFD-314 control group; A2. PDGFD-314-sgRNA2 digestion group; A3. PDGFD-314-sgRNA5 digestion group; B1. PDGFD-410 control group; B2. PDGFD-410-sgRNA digestion group; C1. PDGFD-397 control group; C2. PDGFD-397-sgRNA2 digestion group; C3. PDGFD-397-sgRNA7 digestion group. The arrows indicate the bands produced after digestion"
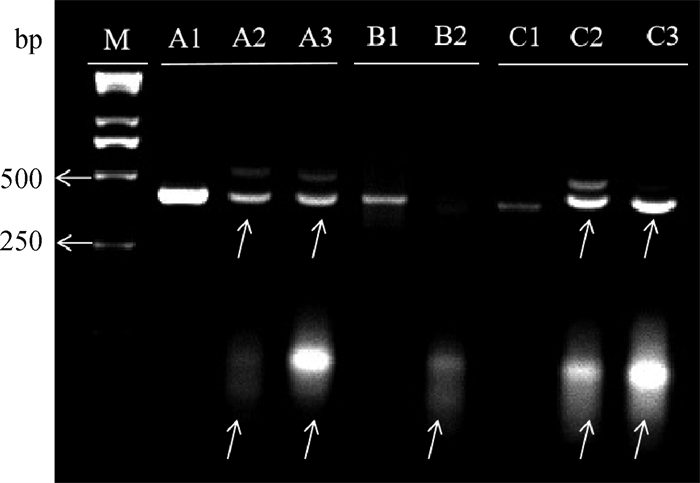
Table 4
Development of electroporated edited zygotes with different editing materials"
| 编辑类型 Editing type | 总卵数/枚 Total number of oocytes | 死亡率/% Mortality rate | 24 h卵裂率/% 24 h cleavage rate | 48 h卵裂率/% 48 h cleavage rate | 7 d囊胚率/% Blastocyst rate on day 7 |
| PDGFD-314-sgRNA2 | 109 | 11.96±0.04 | 46.42±0.12 | 90.37±0.09 | 35.70±0.02B |
| PDGFD-314-sgRNA5 | 107 | 12.22±0.09 | 42.24±0.04 | 89.86±0.10 | 39.92±0.05b |
| PDGFD-410-sgRNA | 109 | 16.98±0.09 | 44.00±0.12 | 93.75±0.08 | 36.77±0.01B |
| PDGFD-397-sgRNA2 | 112 | 19.03±0.09 | 42.60±0.13 | 90.33±0.13 | 32.12±0.10B |
| PDGFD-397-sgRNA7 | 115 | 10.95±0.06 | 45.55±0.12 | 90.69±0.03 | 36.20±0.01B |
| 对照组Control | 118 | 1.60±0.03 | 49.89±0.02 | 79.34±0.08 | 57.73±0.05Aa |
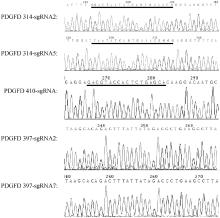
Fig. 2
Amplification and sequencing results of electroporated edited zygotes Sanger sequencing maps of PDGFD-314-sgRNA2, PDGFD-314-sgRNA5, PDGFD-410-sgRNA, PDGFD-397-sgRNA2, and PDGFD-397-sgRNA7 edited embryos. Where the black underline indicates the sgRNA binding site at this site, and the ununderlined indicates that the sgRNA has been deleted at this site"
Table 5
Effects of electroporation editing of different types of genes on embryonic development"
| 编辑类型 Editing type | 总卵数/枚 Total number of oocytes | 死亡率/% Mortality rate | 24 h卵裂率/% 24 h cleavage rate | 48 h卵裂率/% 48 h cleavage rate | 7 d囊胚率/% Blastocyst rate on day 7 |
| PDGFD-397-sgRNA2电转编辑组 PDGFD-397-sgRNA2 electrotransfer editing group | 131.00 | 7.36±0.08 | 54.31±0.91 | 94.26±0.06 | 29.68±0.00b |
| MSTN电转编辑组 MSTN electrotransfer editing group | 120.00 | 11.79±0.06 | 60.67±0.14 | 92.81±0.02 | 51.99±0.15a |
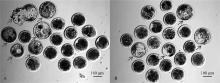
Fig. 3
Representative pictures of sheep blastocyst development on day 7 in PDGFD-397 electroporation editing group and MSTN gene electroporation editing group A. Blastocyst development diagram of MSTN gene electroporation editing group on day 7; B. Blastocyst development diagram of PDGFD-397 electroporation editing group on day 7. The arrows are the blastocysts"
| 1 | FARHADI S , HASANPUR K , GHIAS J S , et al. Comprehensive gene expression profiling analysis of adipose tissue in male individuals from fat-and thin tailed sheep breeds[J]. Animals (Basel), 2023, 13 (22): 3475. |
| 2 |
张越, 曹贵方. 绵羊尾脂沉积的研究进展[J]. 当代畜禽养殖业, 2022 (4): 16- 18.
doi: 10.3969/j.issn.1005-5959.2022.04.006 |
|
ZHANG Y , CAO G F . Research progress on tail fat deposition in sheep[J]. Journal of Contemporary Livestock and Poultry Industry, 2022 (4): 16- 18.
doi: 10.3969/j.issn.1005-5959.2022.04.006 |
|
| 3 | 甘尚权, 张伟, 沈敏, 等. 绵羊X染色体59578440位点多态分析及其与尾(臀)脂性状相关性研究[J]. 新疆农业科学, 2013, 50 (12): 2311- 2316. |
| GAN S Q , ZHANG W , SHEN M , et al. Polymorphism analysis of 59578440 locus on X chromosome of sheep and its correlation with tail (hip) fat traits[J]. Xinjiang Agricultural Sciences, 2013, 50 (12): 2311- 2316. | |
| 4 | 马林. 绵羊尾脂沉积相关lncRNA的比较转录组研究[D]. 杨凌: 西北农林科技大学, 2018. |
| MA L. Comparative transcriptome study of lncRNA related to tail fat deposition in sheep[D]. Yangling: Northwest A&F University, 2018. (in Chinese) | |
| 5 |
宋淑珍, 刘俊斌, 朱才业, 等. 断尾对兰州大尾羊生长性能、脂肪沉积分布和屠宰性能的影响[J]. 畜牧兽医学报, 2023, 54 (2): 642- 655.
doi: 10.11843/j.issn.0366-6964.2023.02.021 |
|
SONG S Z , LIU J B , ZHU C Y , et al. Effects of tail docking on growth performance, fat deposition distribution and slaughter performance of Lanzhou fat-tailed sheep[J]. Acta Veterinaria et Zootechnica Sinica, 2023, 54 (2): 642- 655.
doi: 10.11843/j.issn.0366-6964.2023.02.021 |
|
| 6 |
孟秋赤, 卢光玉, 陈定双, 等. 山羊GPR35基因表达特性分析及对皮下脂肪细胞分化作用的研究[J]. 畜牧兽医学报, 2023, 54 (12): 4993- 5007.
doi: 10.11843/j.issn.0366-6964.2023.12.011 |
|
MENG Q C , LU G Y , CHEN D S , et al. Expression characteristics of GPR35 gene in goat and its effect on subcutaneous adipocyte differentiation[J]. Acta Veterinaria et Zootechnica Sinica, 2023, 54 (12): 4993- 5007.
doi: 10.11843/j.issn.0366-6964.2023.12.011 |
|
| 7 |
ROSEN E D , MACDOGALD O A . Adipocyte differentiation from the inside out[J]. Nat Rev Mol Cell Biol, 2006, 7 (12): 885- 896.
doi: 10.1038/nrm2066 |
| 8 |
梁慧丽, 解玉静, 司博文, 等. 基于全基因组重测序分析大尾寒羊基因组变异特征和群体结构[J]. 畜牧兽医学报, 2024, 55 (11): 4968- 4979.
doi: 10.11843/j.issn.0366-6964.2024.11.016 |
|
LIANG H L , XIE Y J , SI B W , et al. Analysis of genome variation and population structure of large tail Han sheep based on whole genome resequencing[J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55 (11): 4968- 4979.
doi: 10.11843/j.issn.0366-6964.2024.11.016 |
|
| 9 | 王铁男, 刘莉. 羊尾脂的研究及利用现状[J]. 草食家畜, 2021 (3): 6- 10. |
| WANG T N , LIU L . Research and utilization status of sheep tail fat[J]. Herbivorous Livestock, 2021 (3): 6- 10. | |
| 10 | 马丽娜, 马青, 李颖康. 滩羊尾脂组织候选基因mRNA表达量分析研究[J]. 畜牧与饲料科学, 2018, 39 (9): 19- 23. |
| MA L N , MA Q , LI Y K . Analysis of mRNA expression levels of candidate genes in Tan sheep tail fat tissue[J]. Animal Husbandry and Feed Science, 2018, 39 (9): 19- 23. | |
| 11 | 李星艳, 王世银, 许瑞霞, 等. 阿勒泰羊CFD基因的克隆及其在不同营养状态阿勒泰羊尾脂中的表达分析[J]. 新疆农业科学, 2016, 53 (6): 1136- 1144. |
| LI X Y , WANG S Y , XU R X , et al. Cloning of CFD gene in Altay sheep and its expression analysis in tail fat of Altay sheep under different nutritional states[J]. Xinjiang Agricultural Sciences, 2016, 53 (6): 1136- 1144. | |
| 12 | 赵伟利. 脂肪组织差异表达基因RETN、CAV1、PLA2G16与阿勒泰羊尾脂沉积代谢关系的研究[D]. 石河子: 石河子大学, 2015. |
| ZHAO W L. Study on the relationship between differentially expressed genes RETN, CAV1, PLA2G16 in adipose tissue and tail fat deposition metabolism of Altay sheep[D]. Shihezi: Shihezi University, 2015. (in Chinese) | |
| 13 | 许瑞霞. 阿勒泰大尾羊FABP4、ADIPOQ和CFD基因分子克隆及其组织差异性表达的研究[D]. 石河子: 石河子大学, 2015. |
| XU R X. Molecular cloning of FABP4, ADIPOQ and CFD genes in Altay fat-tailed sheep and study on their tissue differential expression[D]. Shihezi: Shihezi University, 2015. (in Chinese) | |
| 14 |
LI X , YANG J , SHEN M , et al. Whole-genome resequencing of wild and domestic sheep identifies genes associated with morphological and agronomic traits[J]. Nat Commun, 2020, 11 (1): 2815.
doi: 10.1038/s41467-020-16485-1 |
| 15 |
DONG K , YANG M , HAN J , et al. Genomic analysis of worldwide sheep breeds reveals PDGFD as a major target of fat-tail selection in sheep[J]. BMC Genomics, 2020, 21 (1): 800.
doi: 10.1186/s12864-020-07210-9 |
| 16 |
PAN Z , LI S , LIU Q , et al. Rapid evolution of a retro-transposable hotspot of ovine genome underlies the alteration of BMP2 expression and development of fat tails[J]. BMC Genomics, 2019, 20 (1): 261.
doi: 10.1186/s12864-019-5620-6 |
| 17 |
ZHU C , LI N , CHENG H , et al. Genome wide association study for the identification of genes associated with tail fat deposition in Chinese sheep breeds[J]. Biol Open, 2021, 10 (5): bio054932.
doi: 10.1242/bio.054932 |
| 18 | LI Q , LU Z , JIN M , et al. Verification and Analysis of Sheep Tail Type-Associated PDGF-D Gene Polymorphisms[J]. Animals (Basel), 2020, 10 (1): 89. |
| 19 |
XU Y X , WANG B , JING J N , et al. Whole-body adipose tissue multi-omic analyses in sheep reveal molecular mechanisms underlying local adaptation to extreme environments[J]. Commun Biol, 2023, 6 (1): 159.
doi: 10.1038/s42003-023-04523-9 |
| 20 |
KALDS P , HUANG S , ZHOU S , et al. ABE-induced PDGFD start codon silencing unveils new insights into the genetic architecture of sheep fat tails[J]. J Genet Genomics, 2023, 50 (12): 1022- 1025.
doi: 10.1016/j.jgg.2023.07.008 |
| 21 |
HOSAKA K , WANG C , ZHANG S , et al. Perivascular localized cells commit erythropoiesis in PDGF-B-expressing solid tumors[J]. Cancer Commun (Lond), 2023, 43 (6): 637- 660.
doi: 10.1002/cac2.12423 |
| 22 |
BERNARD M , MENET R , LECORDIER S , et al. Endothelial PDGF-D contributes to neurovascular protection after ischemic stroke by rescuing pericyte functions[J]. Cell Mol Life Sci, 2024, 81 (1): 225.
doi: 10.1007/s00018-024-05244-w |
| 23 | WENG Z , AHMAD A , LI Y , et al. Emerging roles of PDGF-D signaling pathway in tumor development and progression[J]. Biochim Biophys Acta, 2010, 1806 (1): 122- 30. |
| 24 | 刘洁琼. PDGF-D在乳腺癌进展、转移及化疗药物运输中的作用和机制研究[D]. 长沙: 中南大学, 2011. |
| LIU J Q. The role and mechanism of PDGF-D in breast cancer progression, metastasis and chemotherapy drug delivery[D]. Changsha: Central South University, 2011. (in Chinese) | |
| 25 |
JUNG S C , KANG D , KO E A . Roles of PDGF/PDGFR signaling in various organs[J]. Korean J Physiol Pharmacol, 2024,
doi: 10.4196/kjpp.24.309 |
| 26 | 汪立芹, 何宗霖, 林嘉鹏, 等. 萨福克羔羊卵泡诱导发育效果的研究[J]. 中国农业大学学报, 2015, 20 (4): 141- 146. |
| WANG L Q , HE Z L , LIN J P , et al. Study on the effect of follicle-induced development in Saffoke lambs[J]. Journal of China Agricultural University, 2015, 20 (4): 141- 146. | |
| 27 | 王宇辉. PDGFD基因编辑对滩羊胎儿期脂尾发育的影响研究[D]. 杨凌: 西北农林科技大学, 2024. |
| WANG Y H. Effects of PDGFD gene editing on fetal fat tail development of Tan sheep[D]. Yangling: Northwest A&F University, 2024. (in Chinese) | |
| 28 | KALDS T G P. 基于基因组编辑和组学分析的绵羊PDGFD基因功能研究[D]. 杨凌: 西北农林科技大学, 2022. |
| KALDS T G P. Functional study of PDGFD gene in sheep based on genome editing and omics analysis[D]. Yangling: Northwest A&F University, 2022. (in Chinese) | |
| 29 |
LU W , XU P , DENG B , et al. PDGFD switches on stem cell endothelial commitment[J]. Angiogenesis, 2022, 25 (4): 517- 533.
doi: 10.1007/s10456-022-09847-4 |
| 30 | KIM H J, ChENG P, TRAVISANO S, et al. Molecular mechanisms of coronary artery disease risk at the PDGFD locus[DB/OL]. bioRxiv[Preprint]. 2023: 2023.01.26.525789. |
| 31 |
WENG Y , CHEN N , ZHANG R , et al. An integral blood-brain barrier in adulthood relies on microglia-derived PDGFB[J]. Brain Behav Immun, 2024, 115, 705- 717.
doi: 10.1016/j.bbi.2023.11.023 |
| 32 |
CRISPO M , MULET A P , TESSON L , et al. Efficient generation of myostatin knock-out sheep using CRISPR/Cas9 technology and microinjection into zygotes[J]. PLoS One, 2015, 10 (8): e0136690.
doi: 10.1371/journal.pone.0136690 |
| 33 |
GIM G M , KWON D H , EOM K H , et al. Production of MSTN-mutated cattle without exogenous gene integration using CRISPR-Cas9[J]. Biotechnol J, 2022, 17 (7): e2100198.
doi: 10.1002/biot.202100198 |
| 34 |
GUO R , WANG H , MENG C , et al. Efficient and specific generation of MSTN-edited Hu sheep using C-CRISPR[J]. Genes (Basel), 2023, 14 (6): 1216.
doi: 10.3390/genes14061216 |
| 35 |
PI W , FENG G , LIU M , et al. Electroporation delivery of Cas9 sgRNA ribonucleoprotein-mediated genome editing in sheep IVF zygotes[J]. Int J Mol Sci, 2024, 25 (17): 9145.
doi: 10.3390/ijms25179145 |
| 36 |
PAPPANO W N , STEILITZ B M , SCOTT I C , et al. Use of Bmp1/Tll1 doubly homozygous null mice and proteomics to identify and validate in vivo substrates of bone morphogenetic protein 1/tolloid-like metalloproteinases[J]. Mol Cell Biol, 2003, 23 (13): 4428- 4438.
doi: 10.1128/MCB.23.13.4428-4438.2003 |
| 37 |
CHU LF , LENG N , ZHANG J , ET al . Single-cell RNA-seq reveals novel regulators of human embryonic stem cell differentiation to definitive endoderm[J]. Genome Biol, 2016, 17 (1): 173.
doi: 10.1186/s13059-016-1033-x |
| 38 |
CELAURO E , CARRA S , RODRIGUEZ A , et al. Functional analysis of the cfdp1 gene in zebrafish provides evidence for its crucial role in craniofacial development and osteogenesis[J]. Exp Cell Res, 2017, 361 (2): 236- 245.
doi: 10.1016/j.yexcr.2017.10.022 |
| 39 | 孙珂欣. 滩羊PDGFD基因对其尾部脂肪沉积的影响研究[D]. 杨凌: 西北农林科技大学, 2022. |
| SUN K X. Effect of PDGFD gene on tail fat deposition of Tan sheep[D]. Yangling: Northwest A&F University, 2022. (in Chinese) | |
| 40 |
KALDS P , LUO Q , SUN K , et al. Trends towards revealing the genetic architecture of sheep tail patterning: Promising genes and investigatory pathways[J]. Anim Genet, 2021, 52 (6): 799- 812.
doi: 10.1111/age.13133 |
| [1] | XIE Yaru, JIN Haoyan, KONG Chen, CAI Bei, ZHANG Lingkai. Research Progress of CRISPR/Cas9 System in Livestock Germ Cells [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(2): 479-491. |
| [2] | QIU Meiyu, ZHANG Xuemei, ZHANG Ning, LIU Mingjun. Approach and Application of Prime Editing System [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(4): 1345-1355. |
| [3] | LAN Xinrui, ZHAO Baobao, ZHANG Bihan, LIN Xiaoyu, MA Huiming, WANG Yongsheng. Effects of β-sitosterol on Porcine Oocyte Maturation and Embryonic Development in Vitro [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(4): 1629-1637. |
| [4] | WANG Jiali, YANG Fan, SHAO Wenhua, HUANG Mengyao, CAO Weijun, PU Xiuying, ZHANG Wei, ZHENG Haixue. Construction of Tollip Knockout Pig Kidney Cell Line [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(4): 1810-1818. |
| [5] | ZHANG Duo, TENG Man, ZHANG Zhuo, LIU Jinling, ZHENG Luping, GE Siyu, HAN Fang, LUO Qin, CHAI Shujun, ZHAO Dong, YU Zuhua, LUO Jun. Development and Pathogenicity Analysis of a meq-gene-edited Candidate Marek's Disease Vaccine Strain Generated from a Hypervirulent MDV Variant [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(12): 5672-5683. |
| [6] | Xiuhu DING, Zhiping LIN, Fang ZHAO, Kunlin CHEN, Jifeng ZHONG, Yan ZHANG, Yundong GAO, Huixia LI, Huili WANG, Jianli ZHANG, Qiang DING. Highly Efficient BLG Knockout in Bovine Mammary Epithelial Cells by Using CRISPR/Cas9 [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(10): 4475-4488. |
| [7] | ZHANG Chenjian, LI Yinxia, DING Qiang, LIU Weijia, WANG Huili, HE Nan, WU Jiashun, CAO Shaoxian. Efficient Preparation of CRISPR/Cas9-mediated Goat SOCS2 Gene Edited Embryos [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(1): 129-141. |
| [8] | FEI Xiaoyu, SHI Chaoqun, LIU Xueming, SU Feng, JIANG Yunliang. CRISPR/Cas9 System Mediated Gene Modificated MRC1 in PK15 Cells Reduce PCV2 Replication [J]. Acta Veterinaria et Zootechnica Sinica, 2023, 54(3): 934-946. |
| [9] | YANG Xiaogeng, ZHANG Huizhu, LI Jian, XIANG Hua, HE Honghong. Research Progress of the DNA Methylation in Mammalian Oocyte and Early Embryo Development [J]. Acta Veterinaria et Zootechnica Sinica, 2023, 54(2): 443-450. |
| [10] | CHEN Junzhen, QUAN Ran, FU Qiang, GE Lijuan, YUAN Yuanyuan, ZHANG Chengyuan, LI Jianlin, SHI Huijun. Study on the Effect of Heat Shock Protein HSP90B1 on the Replication of Bovine Viral Diarrhea Virus [J]. Acta Veterinaria et Zootechnica Sinica, 2023, 54(2): 683-693. |
| [11] | ZHANG Chenyibo, YU Tong, REN Binbin, ZHENG Ruizhi, ZHU Wenzhi, SU Jianmin. Mechanism of Epigenetic Reprogramming of Early Animal Embryos [J]. Acta Veterinaria et Zootechnica Sinica, 2023, 54(12): 4898-4909. |
| [12] | ZHANG Shuo, ZHOU Yuxiao, WU Haibo, SUO Lun. Dynamics of Gene Editing Consequence Mediated by Long-term CRISPR/Cas9 System [J]. Acta Veterinaria et Zootechnica Sinica, 2023, 54(10): 4196-4208. |
| [13] | DENG Min'er, LI Na, GUO Yaqiong, FENG Yaoyu, XIAO Lihua. Application of CRISPR/Cas9 System on Gene Editing of Parasitic Protozoa [J]. Acta Veterinaria et Zootechnica Sinica, 2023, 54(1): 69-79. |
| [14] | ZHAO Weimin, WANG Huili, CAO Shaoxian, GUO Rihong, WANG Zeping, CHEN Zhe, XU Kui, FU Yanfeng, LI Bixia, REN Shouwen, CHENG Jinhua. The Study of Base Editing of Porcine CD163 Gene [J]. Acta Veterinaria et Zootechnica Sinica, 2022, 53(4): 1041-1050. |
| [15] | LIU Yue, XUE Xianglan, LI Xiaobo, JIANG Lin, PU Yabin, HE Xiaohong, MA Yuehui, ZHAO Qianjun. Research Progress of the Relationship between Chromatin Accessibility and Animal Embryo Development [J]. Acta Veterinaria et Zootechnica Sinica, 2022, 53(3): 680-687. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||