





Acta Veterinaria et Zootechnica Sinica ›› 2024, Vol. 55 ›› Issue (10): 4646-4659.doi: 10.11843/j.issn.0366-6964.2024.10.036
• Basic Veterinary Medicine • Previous Articles Next Articles
Mengli WU1,2( ), Hualin SUN1,2, Jifei YANG1,2, Yaru ZHAO1,2, Guiquan GUAN1,2, Hong YIN1,2,3, Qingli NIU1,2,*(
), Hualin SUN1,2, Jifei YANG1,2, Yaru ZHAO1,2, Guiquan GUAN1,2, Hong YIN1,2,3, Qingli NIU1,2,*( )
)
Received:2023-11-29
Online:2024-10-23
Published:2024-11-04
Contact:
Qingli NIU
E-mail:15238649312@163.com;niuqingli@caas.cn
CLC Number:
Mengli WU, Hualin SUN, Jifei YANG, Yaru ZHAO, Guiquan GUAN, Hong YIN, Qingli NIU. Construction of a Passaged Porcine Alveolar Macrophage Cell Line Stably Expressing the Porcine BRD4-BD1/2 Protein and Its Effects on ASFV Proliferation[J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(10): 4646-4659.

Fig. 1
Screening for BRD4 functional domains affecting ASFV replication The impact of ASFV replication was assessed following transfection with different doses (1, 2, 4 μg) of BRD4 and its various truncations. The results demonstrated that BRD4-BD1/2 notably enhances the replication of ASFV. The specific sizes of BRD4 and truncations are as follows: the full-length BRD4 is approximately 250 ku; BRD4-BD1/2 is around 100 ku; and BRD4-BD1 is about 30 ku. A. Schematic diagram of each structural domain of BRD4; B. ASFV p30 and p72 protein expression levels after dose transfection of BRD4 and its truncates; C. mRNA levels of ASFV B646L gene post-transfection with varying doses of BRD4 and its truncated forms; D. mRNA levels of ASFV CP204L gene after transfection with different doses of BRD4 and its truncated versions. ns. P>0.05, *. P≤0.05 indicates a significant difference between data, **. P≤0.01, ***. P≤0.001, ****. P≤0.000 1"


Fig. 2
Identification of recombinant plasmids by double digestion M. DL2000 DNA marker; 1 Enzymatic products of pLVX-IRES-Puro-3x flag-BRD4-BD1/2 by EcoRⅠ, BamHⅠ. Through the double digestion experiment with EcoRⅠ and BamHⅠ restriction enzymes, we observed bands of approximately 5 000 bp corresponding to the vector and around 2 000 bp for the target gene. These results indicate the successful construction of the lentiviral expression vector pLVX-IRES-Puro-3x Flag-BRD4-BD1/2"


Fig. 3
Stable expression of BRD4-BD1/2 gene in 3D4/21-BRD4-BD1/2 cell line After infection and puromycin selection by the lentiviral packaging system, total protein was extracted from the positive clone cells after continuous passage. Western Blot analysis showed stable expression of BRD4-BD1/2 protein in the passaged 3D4/21-BRD4-BD1/2 cell line"


Fig. 4
Cell viability of the 3D4/21-BRD4-BD1/2 cell line was assessed at various time points using the CCK-8 assay Cellular viability of 3D4/21-BRD4-BD1/2 cells in comparison to 3D4/21-WT cells was evaluated at various time points (12, 24, 36, and 48 hours) and generations (F10, F20, F30)using the CCK-8 assay. The results indicated no significant differences between the cell lines at the time above intervals (ns, P>0.05), suggesting that overexpression of BRD4-BD1/2 does not significantly affect the viability of 3D4/21 cells under the conditions tested. A. CCK-8 detection of 3D4/21-BRD4-BD1/2 cell line activity at different time points; B. CCK-8 detection of 3D4/21-BRD4-BD1/2 cell line activity at different generations"


Fig. 5
Detection of the effects of ASFV-EGFP infection on BRD4-BD1/2-3D4/21 and 3D4/21-WT cell lines by fluorescence microscopy 3D4/21-BRD4-BD1/2 and 3D4/21-WT cells were infected with ASFV-EGFP at varying multiplicities of infection (MOIs) of 0, 0.1, 1, and 2. Fluorescence intensity was monitored at 36 hours post-infection. The results indicated that the 3D4/21-BRD4-BD1/2 cells exhibited increased fluorescence intensity compared to the WT cells, suggesting that the 3D4/21-BRD4-BD1/2 cell line can enhance ASFV replication"


Fig. 6
The impact of two cell lines on ASFV gene transcription and protein expression under various MOI conditions of ASFV infection Both cell types were infected with varying MOIs of ASFV CN/GS/2018 (0, 0.1, 1, 2), and the transcription levels of CP204L, B646L/GAPDH were measured using RT-qPCR, along with the analysis of viral protein expressions p30 and p72 through Western blot. The results revealed that the 3D4/21-BRD4-BD1/2 cell line exhibited significantly higher transcription of CP204L and B646L genes, as well as the expression of p30 and p72 proteins, compared to the 3D4/21-WT cells. A. Expression levels of ASFV proteins p30 and p72 at different MOIs; B. Grayscale analysis of ASFV p30 and p72 protein expression at different MOIs; C. mRNA levels of ASFV B646L gene at different MOIs; D. mRNA levels of ASFV CP204L gene at different MOIs. ns. P>0.05; *. P≤0.05;***. P≤0.001;****. P≤0.000 1"
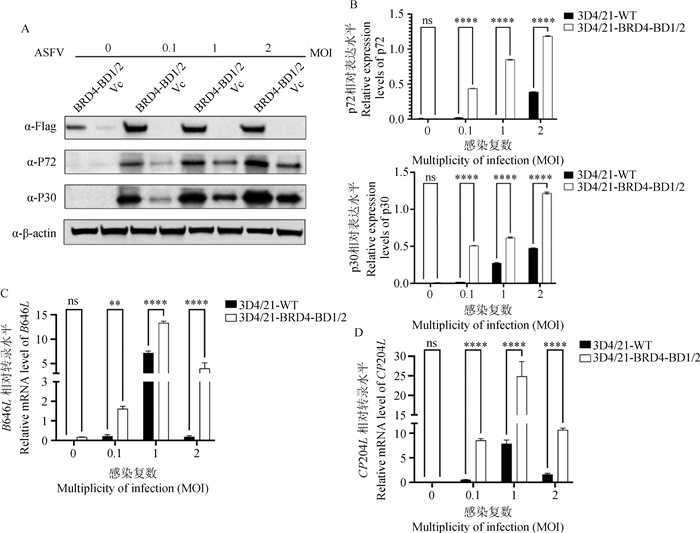

Fig. 7
The influence of two cell lines on ASFV gene transcription and protein expression at different time points during ASFV infection The gene transcription and protein expression profiles at various time points (12, 24, 36, and 48 hpi) post-ASFV CN/GS/2018 infection were analyzed in 3D4/21-BRD4-BD1/2 and 3D4/21-WT cell lines. RT-qPCR technique was utilized to measure the expression levels of CP204L, B646L, and GAPDH mRNA in both cell types. The results indicated a significant increase in the transcription of CP204L and B646L genes at 36 hpi in the 3D4/21-BRD4-BD1/2 cell line (P < 0.01). Additionally, Western blot analysis revealed that the expression of ASFV proteins p30 and p72 was also significantly higher in the 3D4/21-BRD4-BD1/2 cells compared to the 3D4/21-WT cells. A. ASFV p30 and p72 protein expression levels at different time points of infection with ASFV CN/GS/2018; B. Gray value analysis of ASFV p30 and p72 protein expression levels at different time points of infection with ASFV CN/GS/2018; C. mRNA levels of ASFV B646L at different time points; D. mRNA of ASFV CP204L at different time points levels. ns. P > 0.05; *. P≤0.05;***. P≤0.001;****. P≤0.000 1"


Fig. 8
Titer determination of ASFV CN/GS/2018 strain infected 3D4/21 and 3D4/21-BRD4-BD1/2 cells The HAD50 determination method measured the viral titers of ASFV CN/GS/2018 in 3D4/21-BRD4-BD1/2 and 3D4/21-WT cell lines post-infection. The results showed that the HAD50 values for ASFV in the 3D4/21-BRD4-BD1/2 cell line were significantly higher than those in the 3D4/21-WT cell line (P < 0.05). ns. P>0.05; *. P≤0.05;***. P≤0.001;****. P≤0.000 1"


Fig. 9
Analysis of ASFV CN/GS/2018 replication in different generations of 3D4/21-BRD4-BD1/2 cell lines Comparative analysis of ASFV CN/GS/2018 replication capabilities in the 10th, 20th, and 30th generations of 3D4/21-BRD4-BD1/2 cell lines and the control 3D4/21-WT cells. Cells were seeded in 12-well plates and post-infection cell samples were collected at 12, 24, 36, and 48 hours to detect ASFV proteins p30 and p72 using Western blot, indicative of viral replication. The expression of BRD4-BD1/2 in stable cell lines of different generations was examined simultaneously using immunofluorescence. The results from sections A-D indicate that the 3D4/21-BRD4-BD1/2 cell line from the described passages can enhance ASFV replication at the same infection time points compared to WT cells. The results shown in Figure E demonstrate that BRD4-BD1/2 is stably expressed across different passages of the stable expression cell line"

| 1 | 王涛, 孙元, 罗玉子, 等. 非洲猪瘟防控及疫苗研发: 挑战与对策[J]. 生物工程学报, 2018, 34 (12): 1931- 1942. |
| WANG T , SUN Y , LUO Y Z , et al. Prevention, control and vaccine development of African swine fever: challenges and countermeasures[J]. Chinese Journal of Biotechnology, 2018, 34 (12): 1931- 1942. | |
| 2 | 王华, 王君玮, 徐天刚, 等. 非洲猪瘟流行病学和诊断方法的研究进展[J]. 中国兽医科学, 2008, 38 (6): 544- 548. |
| WANG H , WANG J W , XU T G , et al. Advances in studies of epidemiology and diagnostic technologies for African swine fever[J]. Chinese Veterinary Science, 2008, 38 (6): 544- 548. | |
| 3 | 扈荣良, 于婉琪, 陈腾. 非洲猪瘟及防控技术研究现状[J]. 中国兽医学报, 2019, 39 (2): 357- 369. |
| HU R L , YU W Q , CHEN T . African swine fever and its current research status of prevention and control[J]. Chinese Journal of Veterinary Science, 2019, 39 (2): 357- 369. | |
| 4 | 陈腾, 张守峰, 周鑫韬, 等. 我国首次非洲猪瘟疫情的发现和流行分析[J]. 中国兽医学报, 2018, 38 (9): 1831- 1832. |
| CHEN T , ZHANG S F , ZHOU X T , et al. The discovery and epidemic analysis of the first African swine fever epidemic in China[J]. Chinese Journal of Veterinary Science, 2018, 38 (9): 1831- 1832. | |
| 5 |
王清华, 任炜杰, 包静月, 等. 我国首例非洲猪瘟的确诊[J]. 中国动物检疫, 2018, 35 (9): 1- 4.
doi: 10.3969/j.issn.1005-944X.2018.09.001 |
|
WANG Q H , REN W J , BAO J Y , et al. The first outbreak of African swine fever was confirmed in China[J]. China Animal Health Inspection, 2018, 35 (9): 1- 4.
doi: 10.3969/j.issn.1005-944X.2018.09.001 |
|
| 6 | 欧云文, 马小元, 王俊, 等. 非洲猪瘟分子病原学及分子流行病学研究进展[J]. 中国兽医学报, 2018, 38 (2): 416- 420. |
| OU Y W , MA X Y , WANG J , et al. Advances in studies of molecular etiology and molecular epidemiology for African swine fever[J]. Chinese Journal of Veterinary Science, 2018, 38 (2): 416- 420. | |
| 7 |
DIXON L K , CHAPMAN D A G , NETHERTON C L , et al. African swine fever virus replication and genomics[J]. Virus Res, 2013, 173 (1): 3- 14.
doi: 10.1016/j.virusres.2012.10.020 |
| 8 |
BORCA M V , RAI A , RAMIREZ-MEDINA E , et al. A cell culture-adapted vaccine virus against the current African swine fever virus pandemic strain[J]. J Virol, 2021, 95 (14): e0012321.
doi: 10.1128/JVI.00123-21 |
| 9 |
TAKAMATSU H H , DENYER M S , LACASTA A , et al. Cellular immunity in ASFV responses[J]. Virus Res, 2013, 173 (1): 110- 121.
doi: 10.1016/j.virusres.2012.11.009 |
| 10 |
SÁNCHEZ E G , RIERA E , NOGAL M , et al. Phenotyping and susceptibility of established porcine cells lines to African swine fever virus infection and viral production[J]. Sci Rep, 2017, 7 (1): 10369.
doi: 10.1038/s41598-017-09948-x |
| 11 |
ALI H A , LI Y L , BILAL A H M , et al. A comprehensive review of BET protein biochemistry, physiology, and pathological roles[J]. Front Pharmacol, 2022, 13, 818891.
doi: 10.3389/fphar.2022.818891 |
| 12 |
LIANG Y , TIAN J Y , WU T . BRD4 in physiology and pathology: "BET" on its partners[J]. BioEssays, 2021, 43 (12): 2100180.
doi: 10.1002/bies.202100180 |
| 13 | FRANCISCO J C , DAI Q , LUO Z J , et al. Transcriptional elongation control of hepatitis B virus covalently closed circular DNA transcription by super elongation complex and BRD4[J]. Mol Cell Biol, 2017, 37 (19): e00040- 17. |
| 14 | MCBRIDE A A , WARBURTON A , KHURANA S . Multiple roles of Brd4 in the infectious cycle of human papillomaviruses[J]. Front MolBiosci, 2021, 8, 725794. |
| 15 |
ZANDIAN M , CHEN I P , BYRAREDDY S N , et al. Catching BETs by viruses[J]. Biochim Biophys Acta Gene Regul Mech, 2022, 1865 (7): 194859.
doi: 10.1016/j.bbagrm.2022.194859 |
| 16 |
WANG J , LI G L , MING S L , et al. BRD4 inhibition exerts anti-viral activity through DNA damage-dependent innate immune responses[J]. PLoS Pathog, 2020, 16 (3): e1008429.
doi: 10.1371/journal.ppat.1008429 |
| 17 |
GILAN O , RIOJA I , KNEZEVIC K , et al. Selective targeting of BD1 and BD2 of the BET proteins in cancer and immunoinflammation[J]. Science, 2020, 368 (6489): 387- 394.
doi: 10.1126/science.aaz8455 |
| 18 |
刘文豪, 朱彦策, 张冬萱, 等. 稳定表达非洲猪瘟病毒E165R蛋白PK 15细胞系的构建[J]. 畜牧兽医学报, 2023, 54 (6): 2662- 2666.
doi: 10.11843/j.issn.0366-6964.2023.06.042 |
|
LIU W H , ZHU Y C , ZHANG D X , et al. Construction of PK 15 cell line stably expressing African swine fever virus E165R protein[J]. Acta Veterinaria et Zootechnica Sinica, 2023, 54 (6): 2662- 2666.
doi: 10.11843/j.issn.0366-6964.2023.06.042 |
|
| 19 |
张婷, 杨博, 崔卉梅, 等. 过表达非洲猪瘟病毒D1133L蛋白的MA-104细胞系建立及其初步应用[J]. 畜牧兽医学报, 2022, 53 (6): 1877- 1885.
doi: 10.11843/j.issn.0366-6964.2022.06.021 |
|
ZHANG T , YANG B , CUI H M , et al. Establishment and preliminary application of MA-104 cell line overexpressing African swine fever virus D1133L protein[J]. Acta Veterinaria et Zootechnica Sinica, 2022, 53 (6): 1877- 1885.
doi: 10.11843/j.issn.0366-6964.2022.06.021 |
|
| 20 | 殷娟斌, 张志雄, 王莎莎, 等. 过表达HDAC6基因的Vero细胞系建立及其对狂犬病病毒增殖效率评价[J]. 中国兽医科学, 2023, 53 (2): 150- 155. |
| YIN J B , ZHANG Z X , WANG S S , et al. Establishment of Vero cell lines overexpressing HDAC6 gene and evaluation of its proliferative efficiency of rabies virus[J]. Chinese Veterinary Science, 2023, 53 (2): 150- 155. | |
| 21 | RAI A , PRUITT S , RAMIREZ-MEDINA E , et al. Identification of a continuously stable and commercially available cell line for the identification of infectious African swine fever virus in clinical samples[J]. Viruses, 2020, 12 (8): 820. |
| 22 | GAO Y N , XIA T T , BAI J , et al. African swine fever virus exhibits distinct replication defects in different cell types[J]. Viruses, 2022, 14 (12): 2642. |
| 23 | REN K , ZHANG W , CHEN X Q , et al. An epigenetic compound library screen identifies BET inhibitors that promote HSV-1 and -2 replication by bridging P-TEFb to viral gene promoters through BRD4[J]. PLoS Pathog, 2016, 12 (10): e1005950. |
| 24 | TIAN B , LIU Z Q , YANG J , et al. Selective antagonists of the bronchiolar epithelial NF-κB-bromodomain-containing protein 4 pathway in viral-induced airway inflammation[J]. Cell Rep, 2018, 23 (4): 1138- 1151. |
| 25 | HAN X Y , YU D , GU R R , et al. Roles of the BRD4 short isoform in phase separation and active gene transcription[J]. Nat Struct Mol Biol, 2020, 27 (4): 333- 341. |
| 26 | ZHAO Y R , NIU Q L , YANG S X , et al. Inhibition of BET family proteins suppresses African swine fever virus infection[J]. Microbiol Spectr, 2022, 10 (4): e0241921. |
| 27 | JIA N , OU Y W , PEJSAK Z , et al. Roles of African swine fever virus structural proteins in viral infection[J]. J Vet Res, 2017, 61 (2): 135- 143. |
| 28 | TIAN B , YANG J , ZHAO Y X , et al. BRD4 couples NF-κB/RelA with airway inflammation and the IRF-RIG-I amplification loop in respiratory syncytial virus infection[J]. J Virol, 2017, 91 (6): e00007- 17. |
| 29 | RANI A Q , BONAM S R , ZHOU J , et al. BRD4 as a potential target for human papillomaviruses associated cancer[J]. J Med Virol, 2023, 95 (12): e29294. |
| [1] | Lu FENG, Hong TIAN, Haixue ZHENG, Zhengwang SHI, Juncong LUO, Xiaoyang ZHANG, Juanjuan WEI, Jing ZHOU, Huancheng LIAO, Wanying WANG. A Detection Method of African Swine Fever Virus based on Enzymatic Recombinase Amplification [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(9): 4226-4231. |
| [2] | ZHOU Yang, WU Weizi, CAO Weisheng, WANG Fuguang, XU Xiuqiong, ZHONG Wenxia, WU Liyang, YE Jian, LU Shousheng. A Whole Genome Sequencing Method for African Swine Fever Virus based on Nanopore Sequencing Technology was Established [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(5): 2080-2089. |
| [3] | YAN Wenqian, HOU Jing, YANG Jinke, HAO Yu, YANG Xing, SHI Xijuan, ZHANG Dajun, BIE Xintian, CHEN Guohui, CHEN Lingling, HE Lu, ZHAO Meiyu, ZHAO Siyue, ZHENG Haixue, ZHANG Keshan. Monoclonal Antibody against D1133 L Protein of African Swine Fever Virus Inhibits Its Replication [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(2): 854-859. |
| [4] | LIU Chuanxia, WANG Xiao, LI Xuewen, BAO Miaofei, LI Tingting, CHEN Xin, WENG Changjiang, ZHENG Jun. Preparation of Monoclonal Antibody of African Swine Fever Virus pE120R [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(1): 388-394. |
| [5] | FENG Yongzhi, GONG Ting, WU Dongdong, GAO Qi, ZHENG Xiaoyu, ZHANG Guihong, SUN Yankuo. Analysis of Factors Affecting the Infectivity of African Swine Fever Virus on Cultured Cells [J]. Acta Veterinaria et Zootechnica Sinica, 2023, 54(8): 3406-3414. |
| [6] | LIU Taoxue, SU Bingqian, QI Yanli, GUO Jiangtao, LIU Zhonghu, CHU Beibei, WANG Jiang, ZENG Lei. Preparation of the Monoclonal Antibody against the African Swine Fever Virus p30 Protein and Identification of the Antigenic Epitope [J]. Acta Veterinaria et Zootechnica Sinica, 2023, 54(8): 3415-3423. |
| [7] | DING Xiaoyan, HE Jiuxiang, ZHOU Xiaoyang, ZHOU Yuxin, LI Jintao. Preliminary Identification of Host Regulatory Genes and Virulence Genes during African Swine Fever Virus Infection [J]. Acta Veterinaria et Zootechnica Sinica, 2023, 54(7): 2964-2971. |
| [8] | WANG Ying, ZHU Jiahong, ZHAO Jiakai, JI Pinpin, CHEN Xu, ZHANG Lu, LIU Baoyuan, SUN Yani, ZHAO Qin. Screening and Identification of Nanobodies against NP419L Protein of African Swine Fever Virus and Its Preliminary Application of Antibody Detection [J]. Acta Veterinaria et Zootechnica Sinica, 2023, 54(6): 2509-2520. |
| [9] | LIU Wenhao, ZHU Yance, ZHANG Dongxuan, WANG Zhihao, ZHANG Chao. Construction of PK 15 Cell Line Stably Expressing African Swine Fever Virus E165R Protein [J]. Acta Veterinaria et Zootechnica Sinica, 2023, 54(6): 2662-2666. |
| [10] | WANG Guochao, ZHAO Yaru, ZHANG Zhonghui, ZHANG Yulong, BAI Ge, GENG Shuxian, FAN Jie, YANG Jifei, GUAN Guiquan, YIN Hong, LUO Jianxun, NIU Qingli. Bioinformatics Analysis of RNA Polymerase Subunit D205R Gene of African Swine Fever Virus and Polyclonal Antibody Preparation [J]. Acta Veterinaria et Zootechnica Sinica, 2023, 54(5): 2042-2049. |
| [11] | ZHANG Ting, FENG Tao, YANG Jinke, HAO Yu, YANG Xing, ZHANG Dajun, SHI Xijuan, YAN Wenqian, CHEN Lingling, LIU Xiangtao, ZHENG Haixue, ZHANG Keshan. Construction and Growth Characteristics of Recombinant African Swine Fever Virus with Conditional Deletion of D1133L Gene [J]. Acta Veterinaria et Zootechnica Sinica, 2023, 54(2): 706-714. |
| [12] | ZHANG Fangyuan, YANG Dawei, QIU Deyang, JIANG Guoqian, LI Guimei, SHAN Hu. Expression of ASFV P30 Protein and Development of ASFV Antibody Detection Method Based on x-MAP Technology [J]. Acta Veterinaria et Zootechnica Sinica, 2023, 54(10): 4300-4310. |
| [13] | QI Yanli, LIU Taoxue, YU Haishen, ZHANG Chao, LU Weifei, WANG Jiang, CHU Beibei, ZHANG Gaiping. Preparation of the Monoclonal Antibody against the African Swine Fever Virus p54 Protein and Identification of the Antigenic Epitope [J]. Acta Veterinaria et Zootechnica Sinica, 2023, 54(1): 281-292. |
| [14] | WANG Yang, CUI Shuai, XIN Ting, WANG Xixi, YU Hainan, CHEN Shiyu, JIANG Yajun, GAO Xintao, PANG Zhongbao, JIANG Yitong, GUO Xiaoyu, JIA Hong, ZHU Hongfei. ASFV MGF360-14L Interacts with MAVS and Inhibit the Expression of Type Ⅰ Interferon [J]. Acta Veterinaria et Zootechnica Sinica, 2022, 53(9): 3272-3278. |
| [15] | YUAN Xingguo, SHEN Chaochao, ZHAO Dengshuai, YANG Bo, CUI Huimei, HAO Yu, YANG Jinke, CHEN Xuehui, ZHANG Ting, ZHANG Keshan, LIU Xia, ZHENG Haixue, LIU Xiangtao. β-estradiol and Progesterone Promote Replication of African Swine Fever Virus in vitro [J]. Acta Veterinaria et Zootechnica Sinica, 2022, 53(7): 2252-2259. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||