





Acta Veterinaria et Zootechnica Sinica ›› 2025, Vol. 56 ›› Issue (5): 2325-2339.doi: 10.11843/j.issn.0366-6964.2025.05.030
• Preventive Veterinary Medicine • Previous Articles Next Articles
QIAO Yarui1,2( ), MIAO Yuhang1,2, HUANG Qian1,2, ZHOU Xuezhang1,2,*(
), MIAO Yuhang1,2, HUANG Qian1,2, ZHOU Xuezhang1,2,*( )
)
Received:2024-06-17
Online:2025-05-23
Published:2025-05-27
Contact:
ZHOU Xuezhang
E-mail:15209665024@163.com;zhouxuezhang@nxu.edu.cn
CLC Number:
QIAO Yarui, MIAO Yuhang, HUANG Qian, ZHOU Xuezhang. Research on the Biological Characteristics of Enterococcus faecalis in Dairy Cow Mastitis in Ningxia[J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(5): 2325-2339.
Table 1
PCR reaction amplification primers for enterococcal"
| 基因种类 Type of genes | 基因 Genes | 引物序列(5′→ 3′) Sequence | 扩增长度/bp Fragments size | 退火温度/℃ Annealing temperature | 参考文献 Reference |
| 耐药基因 Drug resistance gene | tetM | F-GTGGACAAAGGTACAACGAG | 406 | 56 | [ |
| R-CGGTAAAGTTCGTCACACAC | |||||
| tetA | F-GCTACATCCTGCTTGCCTTC | 210 | 64 | ||
| R-CATAGATCGCCGTGAAGAGG | |||||
| tetB | F-GCCCAGTGCTGTTGTTGTCAT | 617 | 56 | ||
| R-CGTTTTTTCGCCCCATTTAGT | |||||
| tetC | F-CTTGAGAGCCTTCAACCCAG | 418 | 55 | ||
| R-ATGGTCGTCATCTACCTGCC | |||||
| tetX4 | F-CTGATTCGTGTGACATCATCTTTTG | 204 | 56 | ||
| R-GTTAAATTTCCCATTGGTCAGATTA | |||||
| tetX5 | F-GGTATCAACATTTCAATGCTTG | 265 | 56 | ||
| R-CGATTCGTCCTGCGTATCTTTTG | |||||
| mecA | F-TCCAGATTACAACTTCACCAGG | 162 | 54 | ||
| R-CCACTTCATATCTTGTAACG | |||||
| ermA | F-GGATCAGGAAAAGGACATTTTAC | 412 | 54 | ||
| R-GTTCAAGAACAATCAATACAGAG | |||||
| ermB | F-GAAATTGGAACAGGTAAAGGGC | 359 | 56 | ||
| R-GAATCGAGACTTGAGTGTGC | |||||
| ermC | F-GGATCAGGAAAAGGACATTTTAC | 572 | 55 | ||
| R-GCTAATATTGTTTAAATCGTCAATTCC | |||||
| lsaA | F-GGCAATCGCTTGTGTTTTAGCG | 1 200 | 61 | [ | |
| R-GTGAATCCCATGATGTTGATACC | |||||
| lsaE | F-TGTCAAATGGTGAGCAAACG | 496 | 54 | [ | |
| R-TGTAAAACGGCTTCCTGATG | |||||
| lnuA | F-GGTGGCTGGGGGGTAGATGTATTAACTGG | 323 | 54 | [ | |
| R-GCTCTCTTTGAAATACATGGTATTTTTCGATC | |||||
| lnuG | F-AAGGAACTAATGGCAAGGGT | 706 | 53 | [ | |
| R-CCATAACGAGAAGCAAAGAG | |||||
| 毒力基因 Virulence genes | cylA | F-GACTCGGGGATTGATAGGC | 688 | 56 | [ |
| R-GCTGCTAAAGCTGCGCTTAC | |||||
| esp | F-TTGCTAATGCTAGTCCACGACC | 932 | 56 | ||
| R-GCGTCAACACTTGCATTGCCGA | |||||
| asa1 | F-CCAGCCAACTATGGCGGAATC | 529 | 56 | ||
| R-CCTGTCGCAAGATCGACTGTA | |||||
| efaA | F-GCCAATTGGGACAGACCCTC | 688 | 58 | ||
| R-CGCCTTCTGTTCCTTCTTTGGC | |||||
| gelE | F-ACCCCGTATCATTGGTTT | 405 | 56 | ||
| R-ACGCATTGCTTTTCCATC | |||||
| ace | F-GGAATGACCGAGAACGATGGC | 616 | 56 | ||
| R-GCTTGATGTTGGCCTGCTTCCG | |||||
| agg | F- AAGAAAAAGAAGTAGACCAAC | 1 553 | 58 | ||
| R-AAACGGCAAGACAAGTAAATA | |||||
| hyl | F-ACAGAAGAGCAGGAAATG | 276 | 56 | ||
| R-GACTGACGTCCAAGTTTCCAA |
Table 2
7 pairs of primers for housekeeping genes for E. faecalis MLST typing"
| 基因 Genes | 引物序列(5′→ 3′) Sequence | 扩增长度/bp Product size | 退火温度/℃ Annealing temperatures |
| adk | F-GAACCTCATTTTAATGGGG | 437 | 50 |
| R-TGATGTTGATAGCCAGACG | |||
| atpA | F-CGGTTCATACGGAATGGCACA | 556 | |
| R-AAGTTCACGATAAGC CACGG | |||
| ddl | F-GAGACATTGAATATGCCTTATG | 465 | |
| R-AAAAAGAAATCGCACCG | |||
| gyd | F-CAAACTGCTTAGCTCCAAGGC | 395 | |
| R-CATTTCGTTGTCATACCAAGC | |||
| gdh | F-GGCGCACTAAAAGATATGGT | 530 | |
| R-CCAAGATTGGGCAACTTCGTC | |||
| purk | F-CAGATTGGCACATTGAAAG | 492 | |
| R-TTCATTCACATATAGCCCG | |||
| pstS | F-TTGAGCCAAGTCGAAGCTGGA | 583 | |
| R-CGTGATCACGTTCTACTTCC |
Table 3
Results of drug susceptibility testing for E. faecalis"
| 抗生素种类 Types of antibiotics | 敏感 Susceptible | 中介 Mediator | 耐药 Resistant | |||||
| 菌株数 Number of strains | 检出率/% Detection rate | 菌株数 Number of strains | 检出率/% Detection rate | 菌株数 Number of strains | 检出率/% Detection rate | |||
| 青霉素 Penicillin | 87 | 81.3 | - | - | 20 | 18.7 | ||
| 庆大霉素 Gentamicin | 77 | 72.0 | 6 | 5.6 | 24 | 22.4 | ||
| 四环素 Tetracycline | 101 | 94.4 | 2 | 1.9 | 4 | 3.7 | ||
| 红霉素 Erythromycin | 98 | 91.6 | 2 | 1.9 | 7 | 6.5 | ||
| 林可霉素 Lincomycin | 107 | 100 | - | - | - | - | ||
| 恩诺沙星 Enrofloxacin | 73 | 68.2 | 8 | 7.5 | 26 | 24.3 | ||
| 头孢唑林 Cephazolin | 81 | 75.7 | 10 | 9.3 | 16 | 15.0 | ||
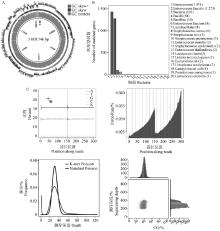
Fig. 1
Information on the bacterial genome(Color images provided in OSID related materials, the same below) A. Bacterial genome circle map; B. Sample gene function annotation NR functional classification map; C. The base content, mass distribution map, 15-mer statistical map, GC content and sequencing depth correlation analysis statistical map of the sample were shown respectively"
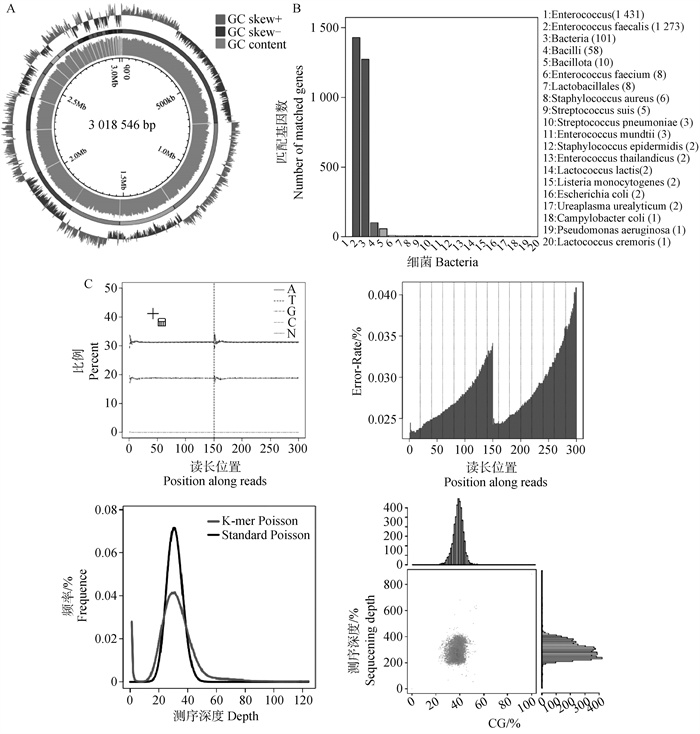
Table 6
Prediction results of resistance gene of E. faecalis genome"
| 菌株 Sample | 抗生素耐药性类型 Type of antibiotic resistance | ||
| F2 | Lincosamide | Macrolide | Gentamincin-b |
| Streptogramin-b | Fluoroquinolone | Amikacin | |
| Spectomycin | Streptomycin | Paromomycin | |
| Tetracycline | Tobramycin | Ribostamycin | |
| Chloramphenicol | Sisomicin | Butirosin | |
| Lincomycin | Dibekacin | Lividomycin | |
| Amikacin | Neomycin | Isepamicin | |
| Netilmicin | Kanamycin | ||
Table 7
Prediction results of virulence factors of E. faecalis genome"
| 菌株 Sample | 毒力因子 Virulence factors | |||
| F2 | DnaK | GAPDH | KatA | MgtBC |
| degP | Ybt | Lap | LysA | |
| SlrA | T6SS-Ⅱ | ClpB | PDH-B | |
| PEB1 | AllB/C/D | YbtQ | SigA | |
| ML1683 | Hyaluronidase | HtrA | Hemolysin Ⅲ | |
| RegX | LPS | VctC/D | UgpC | |
| carB/A | AgrA/C | SrtA | Lsp | |
| PurM | STP | PiuA | HA | |
| GelE | ClpC | Fss1/2 | Lgt | |
| ENO | MprAB | EfaA | TTSS | |
| HemH | Lmb | EF | SprE | |
| EF-Tu | AdsA | ArgK | RelA | |
| GspG | RegX3 | FagC | EPS | |
| SitC | FBPs | Ebp pili | GS | |
| IlpA | AS | CbrC | RmlC | |
| PsaA | FlmH | HitABC | Wzt2 | |
| GlnA1 | LOS | GalE | SSA | |
| Capsule | AI-2 | T4SS | BopD | |
| DltA | Ace | ctpV | FbpABC | |
| LplA1 | VirR/S | AatC | LytR | |
| BSH | Hemolysin | Nuc | GroEL | |
| SodB | NDKs | PgdA | GtcA | |
| RopA | LisR/K | ClpE/P | CdsA | |
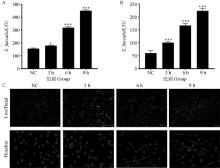
Fig. 7
Toxicity and damage of isolated strains to bovine epithelial cells A.Determination of adhesion ability of bacteria to bovine mammary epithelial cells at different time; B.Determination of invasion ability of bacteria to bovine mammary epithelial cells at different time; C. The effects of bacteria on the damage and proliferation of bovine mammary epithelial cells at different times, in which the red arrow indicates nuclear morphological nuclear pyknosis, cell cavitation, DNA fragments and apoptosis, red indicates dead cells, green indicates living cells. *. P < 0.05, **. P < 0.01, ***. P < 0.001"
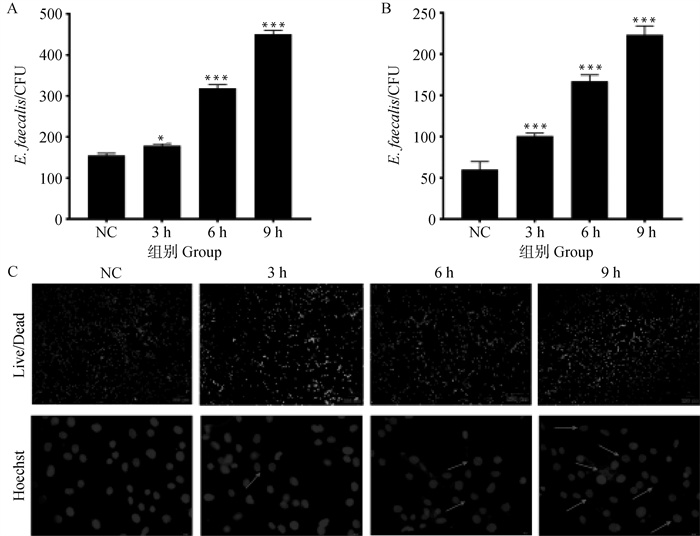
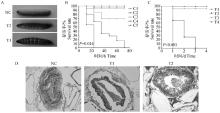
Fig. 8
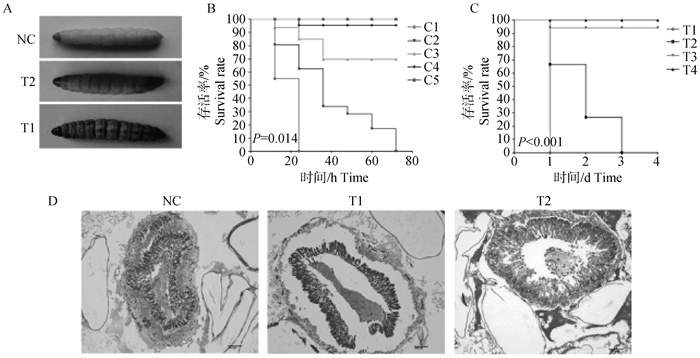
Establishment of animal models and pathogenicity detection of isolated strains A. Changes of body surface color of G. mellonel lalarvae before and after being infected by Enterococci, the body surface became black after infection and death; B. Survival rates of the G. mellonel lalarvae after inoculation with different concentrations of Enterococci [C1. 1×105CFU ·L-1(103/larva); C2. 1×107CFU ·L-1(105/larva); C3. 1×109CFU ·L-1(106/larva); C4. Untreated; C5. Normal saline]; C. Survival rates of the G. mellonel lalarvae after inoculation with different virulence of Enterococci [T1. F2 strain; T2. Negative control; T3. Untreated; T4. Normal saline]; D. Pathological changes of body cavity tissues after the G. mellonel lalarvae were infected by different virulence of Enterococci (HE, 10×)"
| 1 | 白天天,郭雪峰.屎肠球菌的特性及其在畜牧生产中的应用研究进展[J].中国畜牧杂志,2021,57(2):16-20. |
| BAIT T,GUOX F.Characteristics of Enterococcus faecium and its application in livestock production[J].Chinese Journal of Animal Science,2021,57(2):16-20. | |
| 2 | PALMERK L,GILMOREM S.Multidrug-resistant enterococci lack CRISPR-cas[J].mBio,2010,1(4):e00227-10. |
| 3 |
LEBRETONF,MANSONA L,SAAVEDRAJ T,et al.Tracing the enterococci from paleozoic origins to the hospital[J].Cell,2017,169(5):849-861.
doi: 10.1016/j.cell.2017.04.027 |
| 4 |
ZENGJ,TENGF,WEINSTOCKG M,et al.Translocation of Enterococcus faecalis strains across a monolayer of polarized human enterocyte-like T84 cells[J].J Clin Microbiol,2004,42(3):1149-1154.
doi: 10.1128/JCM.42.3.1149-1154.2004 |
| 5 | LEBRETON F, WILLEMS R J L, GILMORE M S. Enterococcus diversity, origins in nature, and gut colonization[M]//GILMORE M S, CLEWELL D B, IKE Y, et al. Enterococci: From commensals to leading causes of drug resistant infection. Boston: Massachusetts Eye and Ear Infirmary, 2014. |
| 6 |
HIRTH,MANIASD A,BRYANE M,et al.Characterization of the pheromone response of the Enterococcus faecalis conjugative plasmid pCF10: complete sequence and comparative analysis of the transcriptional and phenotypic responses of pCF10-containing cells to pheromone induction[J].J Bacteriol,2005,187(3):1044-1054.
doi: 10.1128/JB.187.3.1044-1054.2005 |
| 7 |
LINP Y,CHANS Y,STERNA,et al.Epidemiological profiles and pathogenicity of vancomycin-resistant Enterococcus faecium clinical isolates in Taiwan[J].PeerJ,2023,11,e14859.
doi: 10.7717/peerj.14859 |
| 8 | 胡晓宇,赵才军,曹永国,等.基于肠-乳腺轴浅谈奶牛"内源性乳腺炎"的病理机制及防控措施[J].中国兽医学报,2022,42(6):1287-1294. |
| HUX Y,ZHAOC J,CAOY G,et al.Pathological mechanism and treatment measures ofendogenous mastitis" in dairy cows based on gut-mammarygland axis[J].Chinese Journal of Veterinary Science,2022,42(6):1287-1294. | |
| 9 |
ARIASC A,MURRAYB E.The rise of the Enterococcus: beyond vancomycin resistance[J].Nat Rev Microbiol,2012,10(4):266-278.
doi: 10.1038/nrmicro2761 |
| 10 |
WILLIAMSONA J,JACOBSONR,VAN PRAAGHJ B,et al.Enterococcus faecalis promotes a migratory and invasive phenotype in colon cancer cells[J].Neoplasia,2022,27,100787.
doi: 10.1016/j.neo.2022.100787 |
| 11 |
YANGF,ZHANGS,SHANGX,et al.Short communication: Antimicrobial resistance and virulence genes of Enterococcus faecalis isolated from subclinical bovine mastitis cases in China[J].J Dairy Sci,2019,102(1):140-144.
doi: 10.3168/jds.2018-14576 |
| 12 | 胡晓宇. 奶牛瘤胃菌群紊乱与乳腺炎的关联性及机制研究[D]. 长春: 吉林大学, 2020. |
| HU X Y. The correlation and mechanism between rumen microbiotadisturbance and mastitis in dairy cows[D]. Changchun: Jilin University, 2020. (in Chinese) | |
| 13 |
DE OLIVEIRAR P,ARAGAOB B,DE MELOR,et al.Bovine mastitis in northeastern Brazil: Occurrence of emergent bacteria and their phenotypic and genotypic profile of antimicrobial resistance[J].Comp Immunol Microbiol Infect Dis,2022,85,101802.
doi: 10.1016/j.cimid.2022.101802 |
| 14 |
ROZANSKAH,LEWTAK-PILATA,KUBAJKAM,et al.Occurrence of enterococci in mastitic cow 's milk and their antimicrobial resistance[J].J Vet Res,2019,63(1):93-97.
doi: 10.2478/jvetres-2019-0014 |
| 15 | 曹梦园. 致脑膜炎粪肠球菌突破血脑屏障的相关毒力因子筛选[D]. 石河子: 石河子大学, 2022. |
| CAO M Y. Screening of virulence factors related to blood-brain barrier breakthrough by Enterococeus faecalis[D]. Shihezi: Shihezi University, 2022. (in Chinese) | |
| 16 | ZHANGC,ZHAIS,WUL,et al.Induction of size-dependent breakdown of blood-milk barrier in lactating mice by TiO2 nanoparticles[J].PLoS One,2015,10(4):e122591. |
| 17 | 靳盼盼,刘亚文,邵美丽,等.香芹酚对食源粪肠球菌生物膜形成的抑制作用[J].中国食品学报,2020,20(7):18-26. |
| JINP P,LIUY W,SHAOM L,et al.Inhibition effect of carvacrol to biofilm formation of foodborne Enterococcus faecalis[J].Journal of Chinese Institute of Food Science and Technology,2020,20(07):18-26. | |
| 18 |
GOMEZ-BALTAZARA,VAZQUEZ-GARCIDUENASM S,LARSENJ,et al.Comparative stress response to food preservation conditions of ST19 and ST213 genotypes of Salmonella enterica serotype typhimurium[J].Food Microbiol,2019,82,303-315.
doi: 10.1016/j.fm.2019.03.010 |
| 19 |
LINAG,QUAGLIAA,REVERDYM E,et al.Distribution of genes encoding resistance to macrolides, lincosamides, and streptogramins among staphylococci[J].Antimicrob Agents Chemother,1999,43(5):1062-1066.
doi: 10.1128/AAC.43.5.1062 |
| 20 |
MISICM,KOCICB,ARSOVICA,et al.Human enterococcal isolates as reservoirs for macrolide-lincosamide-streptogramin and other resistance genes[J].J Antibiot (Tokyo),2022,75(7):396-402.
doi: 10.1038/s41429-022-00532-8 |
| 21 | 商艳红. 耐药基因optrA和lsa(E)在猪源肠球菌和链球菌中流行及传播机制的研究[D]. 杨凌: 西北农林科技大学, 2019. |
| SHANG Y H. The epidemiological characteristics and dissemination mechanism of resistance gene optrA and lsa(E) among Enterococcus and Streptococcus suis from swine[D]. Yangling: Northwest A&F University, 2019. (in Chinese) | |
| 22 | 乔亚蕊,杨江流,高登军,等.宁夏地区牛乳肠球菌耐药性分析[J].西北农林科技大学学报(自然科学版),2024,52(8):1-10. |
| QIAOY R,YANGJ L,GAOD Y,et al.Antibiotic resistance of bovine milk derived Enterococcus in Ningxia[J].Journal of Northwest A & F University (Natural Science Edition),2024,52(8):1-10. | |
| 23 |
EATONT J,GASSONM J.Molecular screening of Enterococcus virulence determinants and potential for genetic exchange between food and medical isolates[J].Appl Environ Microbiol,2001,67(4):1628-1635.
doi: 10.1128/AEM.67.4.1628-1635.2001 |
| 24 |
VANKERCKHOVENV,VAN AUTGAERDENT,VAELC,et al.Development of a multiplex PCR for the detection of asa1, gelE, cylA, esp, and hyl genes in enterococci and survey for virulence determinants among European hospital isolates of Enterococcus faecium[J].J CLIN MICROBIOL,2004,42(10):4473-4479.
doi: 10.1128/JCM.42.10.4473-4479.2004 |
| 25 | 何剑琴. 临床感染肠球菌与肠道定植肠球菌的同源性研究[D]. 杭州: 浙江大学, 2021. |
| HE J Q. Homology of Enterococcus from clinical infection to Enterococcus colonization[D]. Hangzhou: Zhejiang University, 2021. (in Chinese) | |
| 26 |
GÖKŞM,TVRK DAǦIH,KARAF,et al.Investigation of antibiotic resistance and virulence factors of Enterococcus faecium and Enterococcus faecalis strains isolated from clinical samples[J].Mikrobiyol Bul,2020,54(1):26-39.
doi: 10.5578/mb.68810 |
| 27 |
胡秀花,孙芷馨,赵梦洋,等.野生松鼠源屎肠球菌的致病性与耐药性分析[J].畜牧兽医学报,2023,54(7):3012-3021.
doi: 10.11843/j.issn.0366-6964.2023.07.032 |
|
HUX H,SUNZ X,ZHAOM Y,et al.Pathogenicity and resistance analysis of Enterococcus faecium from wild squirrels[J].Acta Veterinaria et Zootechnica Sinica,2023,54(7):3012-3021.
doi: 10.11843/j.issn.0366-6964.2023.07.032 |
|
| 28 | CATTOIRV,LECLERCQR.Twenty-five years of shared life with vancomycin-resistant enterococci: is it time to divorce?[J].J Antimicrob Chemother,2013,68(4):731-742. |
| 29 | SHRIDHARP B,AMACHAWADIR G,TOKACHM,et al.Whole genome sequence analyses-based assessment of virulence potential and antimicrobial susceptibilities and resistance of Enterococcus faecium strains isolated from commercial swine and cattle probiotic products[J].J Anim Sci,2022,100(3):skac030. |
| 30 | BAGM A S,ARIFM,RIAZS,et al.Antimicrobial resistance, virulence profiles, and public health significance of Enterococcus faecalis isolated from clinical mastitis of cattle in Bangladesh[J].Biomed Res Int,2022,2022,8101866. |
| 31 | EL-ZAMKANM A,MOHAMEDH.Antimicrobial resistance, virulence genes and biofilm formation in Enterococcus species isolated from milk of sheep and goat with subclinical mastitis[J].PLoS One,2021,16(11):e259584. |
| 32 | TENDOLKARP M,BAGHDAYANA S,GILMOREM S,et al.Enterococcal surface protein, Esp, enhances biofilm formation by Enterococcus faecalis[J].Infect Immun,2004,72(10):6032-6039. |
| 33 | IKEY.Pathogenicity of enterococci[J].Nihon Saikingaku Zasshi,2017,72(2):189-211. |
| 34 | RAHIMIE,SHOKOUHIS,GHAZVINIK,et al.Virulence factors and antimicrobial resistance of Enterococcus faecalis isolated from urinary tract infections[J].J Microbiol, Immunol Infect,2016,49(3):374-379. |
| [1] | WANG Zhuo, ZHAO Yuwei, TU Yan, DIAO Qiyu, CUI Kai. Research Progress on Biological Characteristics of β-defensins and Their Roles in Regulating Intestinal Barrier in Animals [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(3): 995-1005. |
| [2] | YAN Yan, LIU Yanchen, WANG Zhongfa, LI Minjuan, HE Yunan, GUAN Weijun, JIANG Yunliang. Isolation, Culture and Differentiation Potential of Mesenchymal Stem Cells of Yolk Sacs from Rhode Island Red Chicken [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(3): 1252-1263. |
| [3] | Wei LIU, Jiayi MA, Haoyu GENG, Tian XIE, Sunan MIAO, Zongjie LIAO, Shizhong GENG. Isolation, Identification and Characterization of a Broad Spectrum Salmonella Phage [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(9): 4061-4068. |
| [4] | Huanqin ZHENG, Xiaomin JIANG, Hong YUE, Baoyan WANG, Yang LIU, Xingxiao ZHANG, Jianlong ZHANG, Hongwei ZHU. Isolation, Identification and Partial Biological Characteristics Analysis of Feline Herpesvirus-1 [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(7): 3040-3048. |
| [5] | GAO Jie, LI Xiaocheng, MU Yang, ZHANG Hui, WEI Rong, LI Jie. Biological Characteristics and Immune Effect Evaluation of Outer Membrane Vesicles of Capsular Type B Pasteurella multocida [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(5): 2168-2175. |
| [6] | ZHOU Zhiyu, DU Jige, XIN Ruolan, ZHANG Jiawen, PAN Chenfan, YIN Chunsheng, CHEN Xiaoyun, ZHU Zhen. Isolation and Identification of Three Lumpy Skin Disease Viruses in China and Their GPCR Gene Analysis [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(12): 5620-5630. |
| [7] | ZHANG Gaofeng, WEI Jiayang, FENG Helong, LI Li, ZENG Zhe, TIAN Guangming, NIE Renfeng, LUO Qingping, WEN Guoyuan, WEI Hongbo, SHANG Yu. Effects of Biomineralization on the Biological Characteristics and Immunogenicity of the LaSota Strain of Newcastle Disease Virus [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(12): 5663-5671. |
| [8] | Menglei CAI, Dongxu ZHAO, Zhenggang ZHANG, Donghai LIU, Tingting JIANG, Shixuan SU, Xuemin YAN, Xiaoyang XUE, Guolin CUI. The Effect of GreA Protein on the Biological Characteristics and Pathogenicity of Salmonella Enteritidis [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(11): 5173-5182. |
| [9] | LI Zhaoyan, GAO Jiang, GUO Shihui, ZHAO Ruqian, MA Wenqiang. Advances in Detection Methods and Control Solutions for Feline Allergens [J]. Acta Veterinaria et Zootechnica Sinica, 2023, 54(6): 2272-2279. |
| [10] | LIU Qing, WU Shaopeng, SHI Bin, SHAO Hongxia, QIAN Kun, YE Jianqiang, QIN Aijian. Expression of Chicken CTLA-4 Protein in Insect Cells and the Preparation of Its Monoclonal Antibody [J]. Acta Veterinaria et Zootechnica Sinica, 2023, 54(6): 2596-2604. |
| [11] | LI Xiaohan, YANG Fuchun, LIU Rui, GAO Li, CUI Hongyu, ZHANG Yanping, LIU Changjun, QI Xiaole, WANG Xiaomei, GAO Yulong, LI Kai. Construction of Recombinant Herpesvirus of Turkey Expressing Infectious Bursal Disease Virus VP2 Gene and Analysis of Its Biological Characteristics [J]. Acta Veterinaria et Zootechnica Sinica, 2023, 54(2): 656-662. |
| [12] | HAN Shengyi, LI Lingxia, LI Shuping, HU Guoyuan, LI Shengqing. Biological Characteristics and Genome Analysis of Salmonella Phage SP3 [J]. Acta Veterinaria et Zootechnica Sinica, 2023, 54(12): 5228-5239. |
| [13] | CAO Mengyuan, CHEN Mingjie, WANG Chenyu, LI Yitao, YAN Xueqi, CHEN Jie, QI Yayin. Pathological Observation and Transcriptomic Difference Analysis from Mice Infected with Enterococcus faecalis [J]. Acta Veterinaria et Zootechnica Sinica, 2022, 53(9): 3160-3171. |
| [14] | CEN Xin, YANG Tingting, ZHAO Zunfu, WEN Yongping, ZHANG Huanrong. Isolation, Identification, Biological Characteristics and Genome Analysis of a Virulent Salmonella Phage [J]. Acta Veterinaria et Zootechnica Sinica, 2022, 53(8): 2677-2688. |
| [15] | CAO Li, CHENG Runan, WU Zhouhui, WANG Jiawei, WANG Yu, ZHANG Yonghong, WU Qingmin, WANG Zhen. Construction and Biological Characteristics of sapC Gene Deletion Strain of Salmonella Typhimurium [J]. Acta Veterinaria et Zootechnica Sinica, 2022, 53(7): 2282-2289. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||