





Acta Veterinaria et Zootechnica Sinica ›› 2024, Vol. 55 ›› Issue (10): 4346-4359.doi: 10.11843/j.issn.0366-6964.2024.10.010
• Animal Genetics and Breeding • Previous Articles Next Articles
Xiaokun LIN( ), Mengmeng DU, Lisheng ZHOU, Zhengang HUANG, Di WANG, Donghui ZHOU, Xinxin CAO, Jianning HE, Jinshan ZHAO*(
), Mengmeng DU, Lisheng ZHOU, Zhengang HUANG, Di WANG, Donghui ZHOU, Xinxin CAO, Jianning HE, Jinshan ZHAO*( ), Hegang LI*(
), Hegang LI*( )
)
Received:2024-04-15
Online:2024-10-23
Published:2024-11-04
Contact:
Jinshan ZHAO, Hegang LI
E-mail:243252756@qq.com;201501005@qau.edu.cn;201701018@qau.edu.cn
CLC Number:
Xiaokun LIN, Mengmeng DU, Lisheng ZHOU, Zhengang HUANG, Di WANG, Donghui ZHOU, Xinxin CAO, Jianning HE, Jinshan ZHAO, Hegang LI. Genome-Wide Association Study of Wool Economic Traits in Aohan Fine Wool Sheep[J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(10): 4346-4359.
Table 1
Descriptive statistics of wool characteristics of Aohan fine wool sheep"
| 性状 Trait | 平均值 Mean | 标准差 Standard deviation | 最大值 Maximum | 最小值 Minimum | 标准误 Standard error | 变异系数 Coefficient of variation |
| 纤维直径/μm Fiber diameter | 19.633 | 2.248 | 28.124 | 12.935 | 0.124 | 0.115 |
| 自然长度/cm Natural length | 7.643 | 0.562 | 11.758 | 5.913 | 0.031 | 0.074 |
| 伸直长度/cm Elongation length | 10.150 | 0.550 | 4.800 | 7.950 | 0.030 | 0.054 |
| 伸直率/% Elongation rate | 0.331 | 0.061 | 0.612 | 0.167 | 0.003 | 0.183 |

Fig. 5
QQ and Manhattan map of genome-wide association study a. The fiber diameter; b. The natural length; c. The elongation length; d. Elongation rate. In the figure, the horizontal red line represents the genome-wide significance level threshold (P < 4.22×10-5), and the SNP sites above the red line represents the genome-wide significance level. The red line in QQ chart represents the expected value distribution"
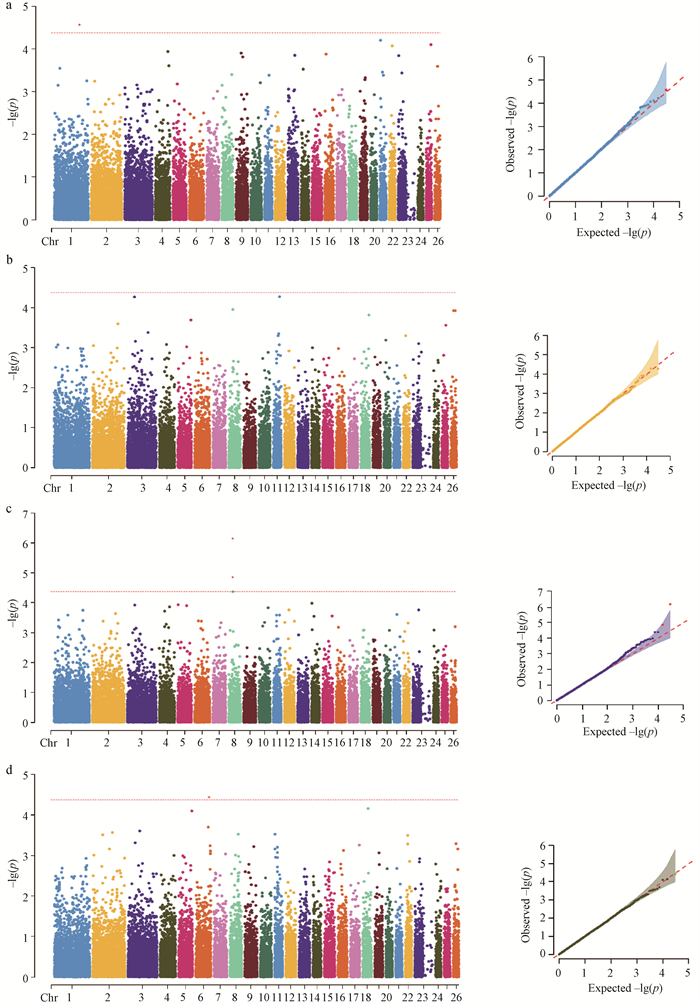
Table 2
Significant SNP sites related to 4 wool traits in sheep"
| 性状Trait | 染色体Chromosome | 位置Location | P值P-value |
| 纤维直径Fiber diameter | 1* | 204762729* | 2.71×10-5* |
| 21 | 6974831 | 6.25×10-5 | |
| 25 | 39039643 | 7.88×10-5 | |
| 22 | 25513088 | 8.52×10-5 | |
| 自然长度Natural length | 11 | 51282011 | 5.29×10-5 |
| 3 | 55043579 | 5.38×10-5 | |
| 伸直长度Elongation length | 8* | 30842589* | 7.05×10-7* |
| 8* | 30837398* | 1.40×10-5* | |
| 8 | 30838595 | 4.26×10-5 | |
| 8 | 30839612 | 4.26×10-5 | |
| 伸直率Elongation rate | 6* | 103863192* | 3.68×10-5* |
| 18 | 41285606 | 6.97×10-5 | |
| 5 | 102267048 | 7.98×10-5 |
Table 3
Information of candidate genes related to 4 wool traits"
| 性状 Trait | 候选基因 Candidate gene | 染色体 Chromosome | 基因位置 Gene location | 基因功能 Gene function |
| 纤维直径 | TTC14 | 1 | 207206481-207216314 | 四肽重复结构域14 |
| Fiber diameter | PEX5L | 1 | 207822971-208123012 | 过氧化物酶体生物发生因子5like |
| SORCS3 | 22 | 24906767-25561739 | sortilin相关VPS10结构域,含受体3 | |
| RAB38 | 21 | 7038110-7103501 | RAB38,RAS癌基因家族成员 | |
| LAMP3 | 1 | 204736807-204768779 | 溶酶体相关膜蛋白3 | |
| MCF2L2 | 1 | 204444714-204732827 | MCF.2细胞系衍生的转化序列样2 | |
| CCSER2 | 25 | 38308913-38471583 | 卷曲螺旋富含丝氨酸的蛋白2 | |
| MCCC1 | 1 | 204782521-204835946 | 甲基巴豆酰辅酶A羧化酶亚基1 | |
| CTSC | 21 | 6903530-6946938 | 组织蛋白酶C | |
| LOC121817525 | 21 | 6905340-6909181 | 未注释 | |
| B3GNT5 | 1 | 204616280-204633302 | UDP-GlcNAc:βGal-β-1, 3-N-乙酰氨基葡萄糖转移酶5 | |
| LOC101103203 | 1 | 204557231-204633105 | LINE-1逆转录转座元件ORF2蛋白 | |
| LOC101114083 | 25 | 38590609-38591984 | 大核糖体亚基蛋白uL4样 | |
| LOC121816833 | 1 | 204742573-204782381 | 未注释 | |
| TRNAG-CCC | 25 | 39009860-39009930 | 转移RNA甘氨酸(反密码子CCC) | |
| TRNAE-UUC | 21 | 6973404-6973477 | 转移RNA谷氨酸(反密码子UUC) | |
| 自然长度 | RPTOR | 11 | 51177578-51498963 | MTOR复合物1的调节相关蛋白 |
| Natural length | ENDOV | 11 | 51502682-51606025 | 核酸内切酶V |
| DNAH6 | 3 | 56369583-56658741 | 动力蛋白轴突重链6 | |
| SUCLG1 | 3 | 51243758-55249958 | 编码异二聚酶琥珀酸辅酶A连接酶的α亚单位 | |
| 伸直长度 | ATG5 | 8 | 30899311-31078799 | 自噬相关5 |
| Elongation length | PRDM1 | 8 | 31122682-31145193 | PR/SET域1 |
| CRYBG1 | 8 | 30674397-30900029 | 含有1个晶体蛋白β-γ结构域 | |
| LOC114116251 | 8 | 30756462-30756568 | U6剪接体RNA | |
| LOC105611045 | 8 | 31060111-31060993 | 小核糖体亚基蛋白eS1样 | |
| LOC114116256 | 8 | 30877462-30877568 | U6剪接体RNA | |
| 伸直率 | MSX1 | 6 | 104950903-104955183 | MSH同源盒1 |
| Elongation rate | TRNAY-GUA | 6 | 105056806-105056878 | 转移RNA酪氨酸(反密码子GUA) |
| ERAP2 | 5 | 93929009-93975030 | 内质网氨肽酶2 | |
| LNPEP | 5 | 93989905-94095313 | 亮氨酰和半胱氨酸氨基肽酶 | |
| NUBPL | 18 | 39830178-40060069 | NUBP铁硫簇组装因子,线粒体 | |
| LOC132658121 | 18 | 40026308-40029156 | 5-羟色胺受体1D样 | |
| NPAS3 | 18 | 41081921..42048771 | 神经元PAS结构域蛋白3 | |
| STX18 | 6 | 105262592-105380010 | 突触融合蛋白18 | |
| ZBTB49 | 6 | 105467844-105494203 | 锌指和BTB结构域含有49 | |
| NSG1 | 6 | 105379653-105413849 | 神经元囊泡运输相关1 | |
| TRNAC-ACA | 6 | 105278250-105278322 | 转移RNA半胱氨酸(反密码子ACA) | |
| TRNAS-GGA | 18 | 41753552-41753623 | 转移RNA丝氨酸(反密码子GGA) | |
| LOC132659842 | 5 | 94094403-94098907 | 未注释 |

Fig. 6
Results of GO enrichment analysis for 39 candidate genes The Y-axis is enrichment terms, and the X-axis is enrichment score. Terms with larger bubbles contain more differential protein-coding genes, and the color of bubbles varies from green to yellow to red. The smaller the P-value, the greater the significance"


Fig. 7
Schematic diagram of candidate genes participating in a common signaling pathway A. Hair follicle development: cell differentiation; B. Autophagy pathway; C. Adaptive immune response partial pathway. The red part is the candidate genes screened in this study; The blue part is a phospholipid compounds"

| 1 |
李莉, 荣威恒, 白俊艳, 等. 影响敖汉细毛羊早期主要性状的非遗传因素分析[J]. 畜牧与饲料科学, 2006, 27 (2): 20- 23.
doi: 10.3969/j.issn.1672-5190.2006.02.012 |
|
LI L , RONG W H , BAI J Y , et al. Analysis of non-genetic factors influencing early main traits in Aohan merino sheep[J]. Animal Husbandry and Feed Science, 2006, 27 (2): 20- 23.
doi: 10.3969/j.issn.1672-5190.2006.02.012 |
|
| 2 |
梅花, 荣威恒. 敖汉细毛羊主要数量性状遗传力的估计[J]. 中国草食动物, 2011, 31 (4): 42- 43.
doi: 10.3969/j.issn.2095-3887.2011.04.013 |
|
MEI H , RONG W H . Estimation of heritability of main quantitative traits in Aohan fine wool sheep[J]. China Herbivores, 2011, 31 (4): 42- 43.
doi: 10.3969/j.issn.2095-3887.2011.04.013 |
|
| 3 |
柳楠, 王春亮, 贺建宁, 等. 敖汉细毛羊不同部位皮肤毛囊发育及形态结构研究[J]. 中国畜牧杂志, 2015, 51 (17): 1- 5.
doi: 10.3969/j.issn.0258-7033.2015.17.001 |
|
LIU N , WANG C L , HE J N , et al. Study on skin hair follicle development and morphology of different body parts of Aohan fine wool sheep[J]. Chinese Journal of Animal Science, 2015, 51 (17): 1- 5.
doi: 10.3969/j.issn.0258-7033.2015.17.001 |
|
| 4 |
WITTENBURG D , BONK S , DOSCHORIS M , et al. Design of experiments for fine-mapping quantitative trait loci in livestock populations[J]. BMC Genet, 2020, 21 (1): 66.
doi: 10.1186/s12863-020-00871-1 |
| 5 |
SHIROKOVA V , BIGGS L C , JUSSILA M , et al. Foxi3 deficiency compromises hair follicle stem cell specification and activation[J]. Stem Cells, 2016, 34 (7): 1896- 1908.
doi: 10.1002/stem.2363 |
| 6 | BOTCHKAREV V A , SHAROV A A . BMP signaling in the control of skin development and hair follicle growth[J]. Differentiation, 2004, 72 (9/10): 512- 526. |
| 7 |
BOLORMAA S , SWAN A A , BROWN D J , et al. Multiple-trait QTL mapping and genomic prediction for wool traits in sheep[J]. Genet Sel Evol, 2017, 49 (1): 62.
doi: 10.1186/s12711-017-0337-y |
| 8 |
WANG C , YUAN Z H , HU R X , et al. Association of SNPs within PTPN3 gene with wool production and growth traits in a dual-purpose sheep population[J]. Anim Biotechnol, 2023, 34 (4): 1429- 1435.
doi: 10.1080/10495398.2022.2029465 |
| 9 |
YUE L , LU Z K , GUO T T , et al. Association of SLIT3 and ZNF280B gene polymorphisms with wool fiber diameter[J]. Animals, 2023, 13 (22): 3552.
doi: 10.3390/ani13223552 |
| 10 |
SALLAM A M , GAD-ALLAH A A , AL-BITAR E M . Association analysis of the ovine KAP6-1 gene and wool traits in Barki sheep[J]. Anim Biotechnol, 2021, 32 (6): 733- 739.
doi: 10.1080/10495398.2020.1749064 |
| 11 |
SALLAM A M , GAD-ALLAH A A , ALBETAR E M . Genetic variation in the ovine KAP22-1 gene and its effect on wool traits in Egyptian sheep[J]. Arch Anim Breed, 2022, 65 (3): 293- 300.
doi: 10.5194/aab-65-293-2022 |
| 12 |
MA G W , CHU Y K , ZHANG W J , et al. Polymorphisms of FST gene and their association with wool quality traits in Chinese Merino sheep[J]. PLoS One, 2017, 12 (4): e0174868.
doi: 10.1371/journal.pone.0174868 |
| 13 |
DARWISH H R , EL-SHORBAGY H M , ABOU-EISHA A M , et al. New polymorphism in the 5' flanking region of IGF-1 gene and its association with wool traits in Egyptian Barki sheep[J]. J Genet Eng Biotechnol, 2017, 15 (2): 437- 441.
doi: 10.1016/j.jgeb.2017.08.001 |
| 14 | KÖCHL S , NIEDERSTÄTTER H , PARSON W . DNA extraction and quantitation of forensic samples using the phenol-chloroform method and real-time PCR[J]. Methods Mol Biol, 2005, 297, 13- 30. |
| 15 | 刘书东. 中国美利奴羊(新疆型)毛品质性状全基因组关联分析[D]. 石河子: 石河子大学, 2017. |
| LIU S D. Genome-wide association study for wool production traits in Chinese Merino sheep (Xinjiang type)[D]. Shihezi: Shihezi University, 2017. (in Chinese) | |
| 16 |
CHANG C C , CHOW C C , TELLIER L C A M , et al. Second-generation PLINK: rising to the challenge of larger and richer datasets[J]. GigaScience, 2015, 4 (1): 7.
doi: 10.1186/s13742-015-0047-8 |
| 17 |
TUERSUNTUOHETI M , ZHANG J H , ZHOU W , et al. Exploring the growth trait molecular markers in two sheep breeds based on Genome-wide association analysis[J]. PLoS One, 2023, 18 (3): e0283383.
doi: 10.1371/journal.pone.0283383 |
| 18 |
YANG J , LEE S H , GODDARD M E , et al. GCTA: a tool for genome-wide complex trait analysis[J]. Am J Hum Genet, 2011, 88 (1): 76- 82.
doi: 10.1016/j.ajhg.2010.11.011 |
| 19 |
LI L , LI Y F , MA Q , et al. Analysis of family structure and paternity test of tan sheep in Yanchi Area, China[J]. Animals, 2022, 12 (22): 3099.
doi: 10.3390/ani12223099 |
| 20 |
ZHANG C , DONG S S , XU J Y , et al. PopLDdecay: a fast and effective tool for linkage disequilibrium decay analysis based on variant call format files[J]. Bioinformatics, 2019, 35 (10): 1786- 1788.
doi: 10.1093/bioinformatics/bty875 |
| 21 |
YU J M , PRESSOIR G , BRIGGS W H , et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness[J]. Nat Genet, 2006, 38 (2): 203- 208.
doi: 10.1038/ng1702 |
| 22 |
ZHOU X , STEPHENS M . Genome-wide efficient mixed-model analysis for association studies[J]. Nat Genet, 2012, 44 (7): 821- 824.
doi: 10.1038/ng.2310 |
| 23 |
PARK M N , CHOI J A , LEE K T , et al. Genome-wide association study of chicken plumage pigmentation[J]. Asian Australas J Anim Sci, 2013, 26 (11): 1523- 1528.
doi: 10.5713/ajas.2013.13413 |
| 24 |
TURNER S D . Qqman: an R package for visualizing GWAS results using Q-Q and manhattan plots[J]. J Open Source Softw, 2018, 3 (25): 731.
doi: 10.21105/joss.00731 |
| 25 |
LI M X , YEUNG J M Y , CHERNY S S , et al. Evaluating the effective numbers of independent tests and significant p-value thresholds in commercial genotyping arrays and public imputation reference datasets[J]. Hum Genet, 2012, 131 (5): 747- 756.
doi: 10.1007/s00439-011-1118-2 |
| 26 |
SAHANA G , GULDBRANDTSEN B , BENDIXEN C , et al. Genome-wide association mapping for female fertility traits in Danish and Swedish Holstein cattle[J]. Anim Genet, 2010, 41 (6): 579- 588.
doi: 10.1111/j.1365-2052.2010.02064.x |
| 27 |
WANG Z P , ZHANG H , YANG H , et al. Genome-wide association study for wool production traits in a Chinese Merino sheep population[J]. PLoS One, 2014, 9 (9): e107101.
doi: 10.1371/journal.pone.0107101 |
| 28 |
RONG E G , YANG H , ZHANG Z W , et al. Association of methionine synthase gene polymorphisms with wool production and quality traits in Chinese Merino population[J]. J Anim Sci, 2015, 93 (10): 4601- 4609.
doi: 10.2527/jas.2015-8963 |
| 29 |
MU F , RONG E G , JING Y , et al. Structural characterization and association of ovine Dickkopf-1 gene with wool production and quality traits in Chinese Merino[J]. Genes (Basel), 2017, 8 (12): 400.
doi: 10.3390/genes8120400 |
| 30 |
LI W R , LIU C X , ZHANG X M , et al. CRISPR/Cas9-mediated loss of FGF5 function increases wool staple length in sheep[J]. FEBS J, 2017, 284 (17): 2764- 2773.
doi: 10.1111/febs.14144 |
| 31 |
ZHAO H C , GUO T T , LU Z K , et al. Genome-wide association studies detects candidate genes for wool traits by re-sequencing in Chinese fine-wool sheep[J]. BMC Genomics, 2021, 22, 127.
doi: 10.1186/s12864-021-07399-3 |
| 32 |
BECKER G M , WOODS J L , SCHAUER C S , et al. Genetic association of wool quality characteristics in United States Rambouillet sheep[J]. Front Genet, 2023, 13, 1081175.
doi: 10.3389/fgene.2022.1081175 |
| 33 |
MARCHINI J , CARDON L R , PHILLIPS M S , et al. The effects of human population structure on large genetic association studies[J]. Nat Genet, 2004, 36 (5): 512- 517.
doi: 10.1038/ng1337 |
| 34 |
VANRADEN P M . Efficient methods to compute genomic predictions[J]. J Dairy Sci, 2008, 91 (11): 4414- 4423.
doi: 10.3168/jds.2007-0980 |
| 35 |
ARMSTRONG R A . When to use the Bonferroni correction[J]. Ophthalmic Physiologic Optic, 2014, 34 (5): 502- 508.
doi: 10.1111/opo.12131 |
| 36 |
WANG J , SHIVAKUMAR S , BARKER K , et al. Comparative study of autoantibody responses between lung adenocarcinoma and benign pulmonary nodules[J]. J Thorac Oncol, 2016, 11 (3): 334- 345.
doi: 10.1016/j.jtho.2015.11.011 |
| 37 | SALOMONIS N, WILLIGHAGEN E, ROUDBARI Z, et al. Ectoderm differentiation (WP2858)[EB/OL]. (2021-05-12). https://pubchem.ncbi.nlm.nih.gov/pathway/WikiPathways:WP2858. |
| 38 | GHOSH M , DENKERT N , REUTER M , et al. Dynamics of the translocation pore of the human peroxisomal protein import machinery[J]. Biol Chem, 2023, 404 (2/3): 169- 178. |
| 39 | LIPIDS C, SLENTER D, WILLIGHAGEN E, et al. Ether lipid biosynthesis (WP5275)[EB/OL]. (2024-01-30). https://www.wikipathways.org/pathways/WP5275.html. |
| 40 | GOMES M A G B , BAUDUIN A , LE ROUX C , et al. Synthesis of ether lipids: natural compounds and analogues[J]. Beilstein J Org Chem, 2023, 19 (1): 1299- 1369. |
| 41 |
KAMRAN M , LAIGHNEACH A , BIBI F , et al. Independent associated SNPs at SORCS3 and its protein interactors for multiple brain-related disorders and traits[J]. Genes, 2023, 14 (2): 482.
doi: 10.3390/genes14020482 |
| 42 |
ZHANG Y Q , LI Y , FAN Y H , et al. SorCS3 promotes the internalization of p75NTR to inhibit GBM progression[J]. Cell Death Dis, 2022, 13 (4): 313.
doi: 10.1038/s41419-022-04753-5 |
| 43 |
BIANCHETTI E , BATES S J , NGUYEN T T T , et al. RAB38 facilitates energy metabolism and counteracts cell death in glioblastoma cells[J]. Cells, 2021, 10 (7): 1643.
doi: 10.3390/cells10071643 |
| 44 |
TANAKA T , WARNER B M , MICHAEL D G , et al. LAMP3 inhibits autophagy and contributes to cell death by lysosomal membrane permeabilization[J]. Autophagy, 2022, 18 (7): 1629- 1647.
doi: 10.1080/15548627.2021.1995150 |
| 45 |
LUNDING L P , KRAUSE D , STICHTENOTH G , et al. LAMP3 deficiency affects surfactant homeostasis in mice[J]. PLoS Genet, 2021, 17 (6): e1009619.
doi: 10.1371/journal.pgen.1009619 |
| 46 |
KANAO H , ENOMOTO T , KIMURA T , et al. Overexpression of LAMP3/TSC403/DC-LAMP promotes metastasis in uterine cervical cancer[J]. Cancer Res, 2005, 65 (19): 8640- 8645.
doi: 10.1158/0008-5472.CAN-04-4112 |
| 47 |
HARALAMBIEVA I H , OBERG A L , DHIMAN N , et al. High-dimensional gene expression profiling studies in high and low responders to primary smallpox vaccination[J]. J Infect Dis, 2012, 206 (10): 1512- 1520.
doi: 10.1093/infdis/jis546 |
| 48 |
MIAO Z R , CAO Q H , LIAO R C , et al. Elevated transcription and glycosylation of B3GNT5 promotes breast cancer aggressiveness[J]. J Exp Clin Cancer Res, 2022, 41 (1): 169.
doi: 10.1186/s13046-022-02375-5 |
| 49 |
SHARLOW E R , LEIMGRUBER S , LIRA A , et al. A small molecule screen exposes mTOR signaling pathway involvement in radiation-induced apoptosis[J]. ACS Chem Biol, 2016, 11 (5): 1428- 1437.
doi: 10.1021/acschembio.5b00909 |
| 50 |
SAXTON R A , SABATINI D M . mTOR signaling in growth, metabolism, and disease[J]. Cell, 2017, 168 (6): 960- 976.
doi: 10.1016/j.cell.2017.02.004 |
| 51 |
SHIRKAVAND A , BOROUJENI Z N , ALEYASIN S A . Examination of methylation changes of VIM, CXCR4, DOK7, and SPDEF genes in peripheral blood DNA in breast cancer patients[J]. Indian J Cancer, 2018, 55 (4): 366- 371.
doi: 10.4103/ijc.IJC_100_18 |
| 52 |
SONG L L , SHEN L J , LI H , et al. Age at natural menopause and hypertension among middle-aged and older Chinese women[J]. J Hypertens, 2018, 36 (3): 594- 600.
doi: 10.1097/HJH.0000000000001585 |
| 53 |
TOMASETTI M , MONACO F , STROGOVETS O , et al. ATG5 as biomarker for early detection of malignant mesothelioma[J]. BMC Res Notes, 2023, 16 (1): 61.
doi: 10.1186/s13104-023-06330-1 |
| 54 |
PASQUALUCCI L , COMPAGNO M , HOULDSWORTH J , et al. Inactivation of the PRDM1/BLIMP1 gene in diffuse large B cell lymphoma[J]. J Exp Med, 2006, 203 (2): 311- 317.
doi: 10.1084/jem.20052204 |
| 55 |
LIU Y Y , LEBOEUF C , SHI J Y , et al. Rituximab plus CHOP (R-CHOP) overcomes PRDM1-associated resistance to chemotherapy in patients with diffuse large B-cell lymphoma[J]. Blood, 2007, 110 (1): 339- 344.
doi: 10.1182/blood-2006-09-049189 |
| 56 |
LI Q , ZHANG L R , YOU W H , et al. PRDM1/BLIMP1 induces cancer immune evasion by modulating the USP22-SPI1-PD-L1 axis in hepatocellular carcinoma cells[J]. Nat Commun, 2022, 13 (1): 7677.
doi: 10.1038/s41467-022-35469-x |
| 57 |
HUANG Y , DE REYNIōS A , DE LEVAL L , et al. Gene expression profiling identifies emerging oncogenic pathways operating in extranodal NK/T-cell lymphoma, nasal type[J]. Blood, 2010, 115 (6): 1226- 1237.
doi: 10.1182/blood-2009-05-221275 |
| 58 |
TAO H Q , GUO L , CHEN L F , et al. MSX1 inhibits cell migration and invasion through regulating the Wnt/β-catenin pathway in glioblastoma[J]. Tumor Biol, 2016, 37 (1): 1097- 1104.
doi: 10.1007/s13277-015-3892-2 |
| 59 |
MATTORRE B , TEDESCHI V , PALDINO G , et al. The emerging multifunctional roles of ERAP1, ERAP2 and IRAP between antigen processing and renin-angiotensin system modulation[J]. Front Immunol, 2022, 13, 1002375.
doi: 10.3389/fimmu.2022.1002375 |
| 60 |
MARUSINA A I , JI-XU A , LE S T , et al. Cell-specific and variant-linked alterations in expression of ERAP1, ERAP2, and LNPEP aminopeptidases in psoriasis[J]. J Invest Dermatol, 2023, 143 (7): 1157- 1167. e10.
doi: 10.1016/j.jid.2023.01.012 |
| 61 |
PALADINI F , FIORILLO M T , TEDESCHI V , et al. The multifaceted nature of aminopeptidases ERAP1, ERAP2, and LNPEP: from evolution to disease[J]. Front Immunol, 2020, 11, 1576.
doi: 10.3389/fimmu.2020.01576 |
| 62 |
ONG J , TIMENS W , RAJENDRAN V , et al. Identification of transforming growth factor-beta-regulated microRNAs and the microRNA-targetomes in primary lung fibroblasts[J]. PLoS One, 2017, 12 (9): e0183815.
doi: 10.1371/journal.pone.0183815 |
| 63 |
TU M S , YIN X Q , ZHUANG W Z , et al. NSG1 promotes glycolytic metabolism to enhance Esophageal squamous cell carcinoma EMT process by upregulating TGF-β[J]. Cell Death Discov, 2023, 9 (1): 391.
doi: 10.1038/s41420-023-01694-6 |
| 64 |
WANG Y H , WU N , SUN D L , et al. NUBPL, a novel metastasis-related gene, promotes colorectal carcinoma cell motility by inducing epithelial-mesenchymal transition[J]. Cancer Sci, 2017, 108 (6): 1169- 1176.
doi: 10.1111/cas.13243 |
| 65 |
GUILLEMYN B , DE SAFFEL H , BEK J W , et al. Syntaxin 18 defects in human and zebrafish unravel key roles in early cartilage and bone development[J]. J Bone Miner Res, 2023, 38 (11): 1718- 1730.
doi: 10.1002/jbmr.4914 |
| 66 | HANSPERS K, WILLIGHAGEN E, WEITZ E. Amino acid metabolism (WP3925)[EB/OL]. (2021-05-16). https://pubchem.ncbi.nlm.nih.gov/pathway/WikiPathways:WP3925. |
| [1] | Hongyan HUANG, Liyun ZHANG, Zhirong HUANG, Zhongping WU, Xumeng ZHANG, Hongjia OUYANG, Junpeng CHEN, Zhenping LIN, Yunbo TIAN, Xiujin LI, Yunmao HUANG. The Study on Population Genetic Diversity and Genome-wide Association Study of Body Weight and Size Traits for Lion-head Geese [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(9): 3914-3924. |
| [2] | Tana AN, Haige HAN, Togtokh MONGKE, Baoyindeligeer MONGKEJARGAL, Wenbo LI, Manglai DUGARJAVIIN. A Review of the Genetic Characteristics of Different Coat Colors in Domestic Horses [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(8): 3297-3308. |
| [3] | Ruiqi ZHANG, Yanqin PANG, Zaishan LI, Xiuguo SHANG, Ganqiu LAN, Jinbiao GUO, Yunxiang ZHAO. Research on Feeding Capacity Selection of Lactating Sows Based on Intelligent Precision Feeding [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(7): 2890-2900. |
| [4] | CUI Shengdi, WANG Kai, ZHAO Zhenjian, CHEN Dong, SHEN Qi, YU Yang, WANG Junge, CHEN Ziyang, YU Shixin, CHEN Jiamiao, WANG Xiangfeng, TANG Guoqing. Identification of Candidate Genes for Pork Texture Traits Using GWAS Combined with Co-localisation of DNA Methylation [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(5): 1945-1957. |
| [5] | TANG Xinxin, ZHENG Jumei, LUO Na, YING Fan, ZHU Dan, LI Sen, LIU Dawei, AN Bingxing, WEN Jie, ZHAO Guiping, LI Hegang. Genetic Mechanism of Broiler Leg Disease Based on Genome-Wide Association Analysis [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(1): 99-109. |
| [6] | LI Keanning, DU Lili, AN Bingxing, DENG Tianyu, LIANG Mang, CAO Sheng, DU Yueying, XU Lingyang, GAO Xue, ZHANG Lupei, LI Junya, GAO Huijiang. Genetic Parameter Estimation and Genome-Wide Association Study for Carcass Traits and Primal Cuts Weight Traits in Huaxi Cattle [J]. Acta Veterinaria et Zootechnica Sinica, 2023, 54(9): 3664-3676. |
| [7] | WANG Zhenyu, ZHANG Saibo, LIU Wenhui, LIANG Dong, REN Xiaoli, YAN Lei, YAN Yuefei, GAO Tengyun, ZHANG Zhen, HUANG Hetian. Genomic Inbreeding Coefficient Analysis and Functional Gene Screening in Different Dairy Farms Based on SNP Chip Data [J]. Acta Veterinaria et Zootechnica Sinica, 2023, 54(7): 2848-2857. |
| [8] | ZHANG Xiaoke, LIAO Weili, CHEN Xinyou, LI Tingting, YUAN Xiaolong, LI Jiaqi, HUANG Xiang, ZHANG Hao. Genome-wide Association Study for Identifying Candidate Genes of Growth Traits in Duroc Pigs [J]. Acta Veterinaria et Zootechnica Sinica, 2023, 54(5): 1868-1876. |
| [9] | FAN Chenyu, SHAN Yanju, ZHANG Ming, JI Gaige, JU Xiaojun, TU Yunjie, HE Xi, SHU Jingting, LIU Yifan, ZHANG Haihan. Genome-wide Association Study of Body Weight and Meat Quality Traits in Lihua Mahuang Chickens [J]. Acta Veterinaria et Zootechnica Sinica, 2023, 54(12): 4982-4992. |
| [10] | ZHANG Changzheng, LI Desen, HUANG Min, FANG Xiaomin, ZHAO Weimin, REN Shouwen, DONG Huansheng, REN Jun, ZHOU Lisheng. An Imputed Whole-genome Sequence-based GWAS Approach Pinpoints Genetic Loci for Body Conformation at Birth and Teat Number Traits in Sushan Pig Population [J]. Acta Veterinaria et Zootechnica Sinica, 2023, 54(1): 88-102. |
| [11] | TAO Wei, HOU Liming, WANG Binbin, LIU Hang, LI Kaijun, YIN Yanzhen, GUO Hao, NIU Peipei, ZHANG Zongping, LI Qiang, HUANG Ruihua, LI Pinghua. Identification of Candidate Genes Affecting Drip Loss in Pork by Genome-wide Selection Signal Method [J]. Acta Veterinaria et Zootechnica Sinica, 2022, 53(5): 1373-1383. |
| [12] | LU Yujie, MO Jiayuan, QI Wenjing, ZHU Siran, YANG Lili, LIU Qiaoling, BU Yage, LAN Ganqiu, LIANG Jing. Population Genetic Structure and Selection Signatures Associated with Litter Size Trait in Several Chinese Indigenous Pig Breeds [J]. Acta Veterinaria et Zootechnica Sinica, 2022, 53(2): 360-369. |
| [13] | LI Hongwei, XU Lingyang, WANG Zezhao, CAI Wentao, ZHU Bo, CHEN Yan, GAO Xue, ZHANG Lupei, GAO Huijiang, LI Junya. Genome-wide Association Study of Slaughter Traits Based on Haplotype in Beef Cattle [J]. Acta Veterinaria et Zootechnica Sinica, 2022, 53(12): 4232-4243. |
| [14] | YANG Xinting, ZHENG Maiqing, TAN Xiaodong, ZHAO Guiping, HUANG Chao, LI Sen, LI Wei, WEN Jie, LIU Ranran. Genetic Parameters Estimation and Key Genes Identification for Meat Quality Traits of Fast-growing Yellow-feather Meat-type Chickens [J]. Acta Veterinaria et Zootechnica Sinica, 2021, 52(9): 2416-2428. |
| [15] | MI Bunong, ZHANG Liguo, BAI Urhan, GUO Yulin, WANG Chunwei, XU Quanzhong, FENG Shuang, LI Guangpeng, SU Xiaohu, ZHANG Li. Genome-wide Association Study of Milk Production Traits in Dairy Meade Sheep [J]. Acta Veterinaria et Zootechnica Sinica, 2021, 52(11): 3294-3303. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||