





Acta Veterinaria et Zootechnica Sinica ›› 2025, Vol. 56 ›› Issue (11): 5335-5351.doi: 10.11843/j.issn.0366-6964.2025.11.001
• Review • Previous Articles Next Articles
YUE Yibing1,2( ), LI Junliang1(
), LI Junliang1( ), BAO Binwu1,3, GAO Chen1, CHEN Yan1, ZHU Bo1, ZHANG Lupei1, WANG Zezhao1, GAO Huijiang1, GAO Xue1,*(
), BAO Binwu1,3, GAO Chen1, CHEN Yan1, ZHU Bo1, ZHANG Lupei1, WANG Zezhao1, GAO Huijiang1, GAO Xue1,*( ), HUANG Yongzhen2,*(
), HUANG Yongzhen2,*( ), LI Junya1,*(
), LI Junya1,*( )
)
Received:2025-04-16
Online:2025-11-23
Published:2025-11-27
Contact:
GAO Xue, HUANG Yongzhen, LI Junya
E-mail:yueyibing2023@163.com;lijunliang@caas.cn;gaoxue@caas.cn;hyzsci@nwafu.edu.cn;lijunya@caas.cn
CLC Number:
YUE Yibing, LI Junliang, BAO Binwu, GAO Chen, CHEN Yan, ZHU Bo, ZHANG Lupei, WANG Zezhao, GAO Huijiang, GAO Xue, HUANG Yongzhen, LI Junya. Research Progress on OMEGA Gene Editing System: Structure, Function, and Optimization Strategies[J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(11): 5335-5351.

Fig. 1
Structure of IscB protein and IscB-ωRNA-DNA ternary complex diagram[17] a. Schematic of IscB protein domains. The IscB protein primarily consists of the N-terminal PLMP domain, RuvC domain, HNH domain, WED domain, and TI domain. The PLMP domain, located at the N-terminus of the protein, is involved in protein localization. The RuvC domain and HNH domain are related to DNA cleavage. The WED domain is mainly involved in binding to both the protein and DNA. The TI domain is related to the stability of the protein. b. IscB-ωRNA-DNA ternary complex diagram. The ωRNA binds to the complementary sequence of DNA, directing the protein to recognize specific DNA sequences. The WED domain forms hydrogen bonds with the single-stranded DNA terminus through specific amino acid residues. The RuvC domain forms direct chemical bonds with the DNA backbone through its catalytic active site, participating in DNA cleavage. TI represents TAM-interacting region. TS represents target strand. NTS represents non-target strand"
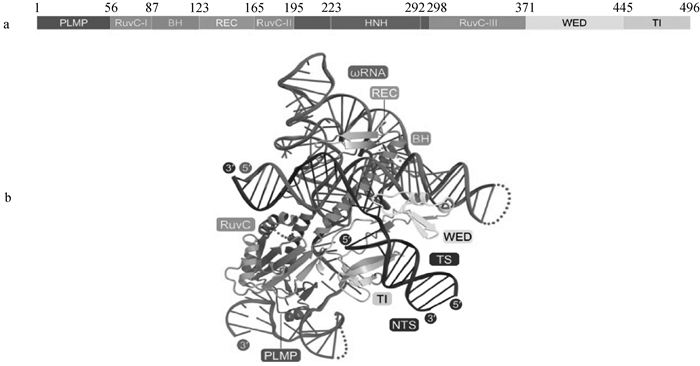
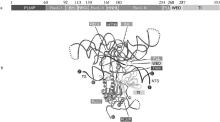
Fig. 2
Structure of IsrB protein and IsrB-ωRNA-DNA ternary complex diagram[23] a. Schematic of IsrB protein domains. The IsrB protein primarily consists of the N-terminal PLMP domain, RuvC domain, WED domain, and TI domain. The PLMP domain, located at the N-terminus of the protein, is involved in protein localization; The WED domain is mainly involved in binding to both the protein and DNA; The RuvC domain is related to DNA cleavage; The TI domain is related to the stability of the protein. b. IsrB-ωRNA-DNA ternary complex diagram. The ωRNA binds to the complementary sequence of DNA, directing the protein to recognize specific DNA sequences; The WED domain forms hydrogen bonds with the single-stranded DNA terminus through specific amino acid residues; The RuvC domain forms direct chemical bonds with the DNA backbone through its catalytic active site, participating in DNA cleavage; PLL represents phosphate-lock loop. TI represents TAM-interacting region. TS represents target strand. NTS represents non-target strand"
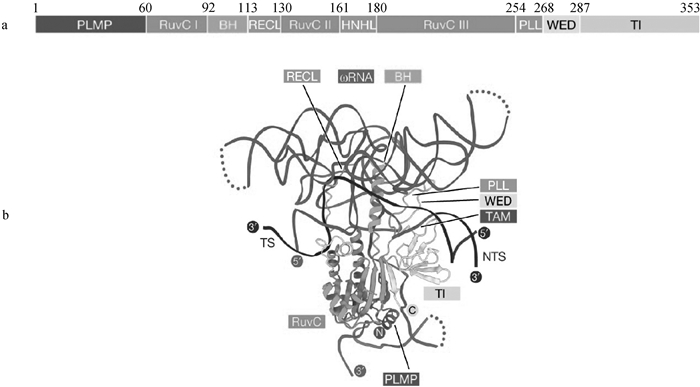
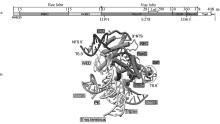
Fig. 3
Structure of TnpB protein and TnpB-reRNA-DNA ternary complex diagram[32] a. Schematic of TnpB protein domains. The TnpB protein consists of an N-terminal recognition (Rec) lobe and a C-terminal nuclease (Nuc) lobe. The Rec lobe includes the REC and wedge (WED) domains. The Nuc lobe includes the RuvC domain and the ZnF domain. The REC domain is mainly responsible for recognizing DNA; The WED domain is primarily involved in binding to the protein and DNA; The RuvC domain is associated with DNA cleavage; The ZnF domain is related to DNA binding. b. Ternary structure of TnpB-reRNA-DNA. The reRNA binds to the complementary sequence of DNA, directing the protein to recognize specific DNA sequences; The WED domain forms hydrogen bonds with the single-stranded DNA terminus through specific amino acid residues; The RuvC domain forms direct chemical bonds with the DNA backbone through its catalytic active site, participating in DNA cleavage; The ZnF domain binds to specific DNA sequences through its zinc finger structure, making the binding more stable; TS represents target strand; NTS represents non-target strand"
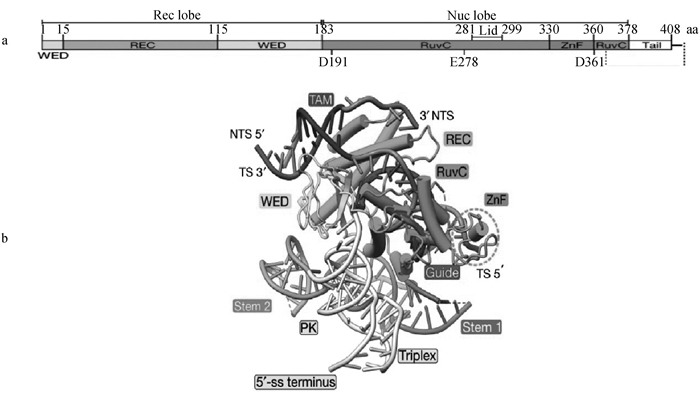
Table 1
Optimization strategies and research progress for the OMEGA gene editing system"
| 时间 Date | 优化系统 Optimization system | 蛋白大小/aa Protein size | ωRNA长度/nt ωRNA length | 编辑对象 Editing target | 优化方案 Optimization strategies | 优化后编辑效率 Optimized editing efficiency | 优化后碱基编辑器编辑效率 Optimized base editor efficiency | 参考文献 Reference | ||
| OMEGA系统核酸酶设计 The OMEGA system nuclease design | ωRNA工程化改造 ωRNA engineering | 融合脱氨酶 Fusion deaminase for base editing | ||||||||
| 2023/5/25 | OgeuIscB-ωRNA系统 | 496 | 222 | HEK293T细胞 | 1.氨基酸替换:E85R、H369R、S387R和S457R得到IscB*;2.融合T5核酸外切酶(T5E):基于enIscB将T5E融合到IscB蛋白C端得到enIscB-T5E | R1茎环截短15 bp(M3)、R2茎环第10碱基对被G-C碱基对独立替换,得到ωRNA* | miABE和miCBE | enIscB-T5E(60.61%±27.43%)在23个目标位点显示出与SpG Cas9(53.25%±30.92%)相当的编辑效率 | miABE编辑效率可达52.37%~60.06%;miCBE编辑效率可达50.17±2.49%~66.42%±1.13%。 | [ |
| 2024/7/8 | OgeuIscB-ωRNA系统 | 496 | 222 | HEK293T、A549 and HeLa细胞 | 氨基酸替换:M102R、F137K、V159K、N281R、Q324R、Y327K、H368R和L393K得到enOgeuIscB | 将茎环中的无序区域使用更稳定的四环“GAAA”取代,并恢复茎环2中的错配,得到ωRNA-v2;在ωRNA-v2的基础上恢复茎环1中的错配得到ωRNA-v13 | enOgeuIscB BE | 在6个基因组位点上观察到64.5%~87.3%的编辑效率 | 优化后的编辑效率达到36.1%±21.1%,野生型OgeuIscB-ωRNA的编辑率为20.3%±21.2% | [ |
| 2024/7/8 | OgeuIscB-ωRNA系统 | 496 | 222 | HEK293T细胞 | 氨基酸替换:D97K、F138N、S431K、S457R得到IscBnQM | 删除了ωRNA的部分茎环1和3′末端生成了优化的ωRNA变体 | SIminiBEs | 与野生型IscBn相比,IscBnQM编辑效率从3.16%~10.13%提高到70.85%~84.60%,优化的ωRNA变体比野生型编辑效率提高了约2倍 | 编辑效率可达50%~60%,提高约2~3倍 | [ |
| 2024/8/15 | IscB.m16系统 | 400左右 | 200~300 | HEK293T细胞 | 氨基酸替换:E326R、P460S、T462H和T459E得到IscB.m16RESH | R1茎环截短13 bp、R5茎环截短10 bp、第24、25、57、79、117和189 bp的碱基对替换为G-C,将其命名为enωRNA | IscB.m16*-ABE和IscB.m16*-CBE | 工程化的IscB.m16*编辑效率在多个位点都超过了40%,比野生型提高2~9倍 | IscB.m16*-ABE(46.15%±4.08%)显示出显著高于IscB.m16-ABE(9.19%± 2.34%);IscB.m16*-CBE编辑效率显著高于SpG-CBE | [ |
| 2024/8/22 | OgeuIscB-ωRNA系统 | 496 | 222 | HEK293T细胞 | 1.氨基酸替换:D96R、E84R、V159R,融合HMG-D结构域;得到IscB-DIscB-D2.核定位信号优化:进行了SV40 NLS融合,优化后的变体被命名为eIscB-D | P1茎环截短15 bp、P2茎环截短4 bp、P5茎环截短4 bp、末端发夹截断、在第11位用G-C配对替换A-U配对和在第103位用A替换C以形成A-U配对,命名为eωRNA | eiABE,eiCBE | eIscB-D编辑效率最高可达91.3%;与原始IscB相比,工程化的eIscB-D/eωRNA系统的活性平均增加了20.2倍 | eiABE效率高达58.3%,eiCBE效率为35.7%~79.2%(平均61.9%) | [ |
| 2024/11/26 | enIscB系统 | 496 | 163 | HEK293T细胞 | 无 | 茎环2的完全去除、茎环3中88号位置的A: U配对替换以及3′尾序列的14个核苷酸缺失,最终长度为163 nt,命名为ωRNA*-v2 | 对miABE、miCBE进行AAV包装,在酪氨酸血症小鼠模型中实现了疾病纠正 | 编辑效率可达87.3% | 碱基编辑效率可达62.2% | [ |
| 2024/1/27 | ISDra2 TnpB系统 | 408 | 231 | HEK293T、小鼠N2a细胞、植物细胞(nicotiana benthamiana,本氏烟草) | 无 | S1、S2和S3的同时缺失并用5′-GAAA-3′环序列替换SL3亚结构域的序列得到ωRNA*变体(99nt) | 无 | 与野生型ωRNA相比,ωRNA*的基因编辑效率提高了两倍(70%左右),与SaCas9效率相当 | 无 | [ |
| 2024/3/19 | ISDra2 TnpB系统 | 408 | 231 | HEK293T和HeLa细胞,小鼠NIH/3T3细胞 | 蛋白结构域截短:截短CTD结构域,TnpB379 | 无 | 无 | TnpB379(29.0%±12.3%)编辑效率高于TnpB(26.4%±10.7%) | 无 | [ |
| 2024/6/28 | ISDra2 TnpB系统 | 408 | 231 | 水稻、拟南芥 | 1.密码子优化:将ISDra2 TnpB基因的密码子替换为植物细胞偏好的密码子;2.启动子优化:使用Pol-II启动子(ZmUbi)替代Pol-III(RNA聚合酶Ⅲ)启动子(OsU3)得到TnpB2 | 无 | D191A突变,得到TnpB-ABE | 在植物基因组中实现了平均高达33.58% 的编辑效率 | 编辑效率只有0.42%~1.12% | [ |
| 2024/8/7 | TnpB系统 | 408 | 231 | 水稻、拟南芥、青蒿、丹参、黄芩、靛蓝和党参 | 1.核定位信号优化:TnpB载体的两个末端融合了真核核定位信号,将TnpB与黄色荧光蛋白(YFP)融合生成了YFP-TnpB构建体;2.启动子优化:由U6和UBQ1启动子驱动reRNA和TnpB载体 | 无 | 无 | 编辑效率最高达30% | 无 | [ |
| 2024/8/21 | IsDge10 TnpB系统 | 400左右 | 200~300 | 水稻 | 启动子优化:IsDge10蛋白在ZmUbi1启动子下表达,7个sgRNA在OsUbi启动子下表达并融合了HH-HDV双核酶系统 | 无 | 无 | 在水稻原生质体中的效率为4.3% 至18.2% | 无 | [ |
| 2024/9/23 | ISDra2 TnpB系统 | 408 | 231 | HEK293T细胞、小鼠肝脏、大脑 | 1.氨基酸替换:蛋白第76位引入氨基酸替换(K76A、K76C和K76S);2.核定位信号优化:ARC13设计(在C端包含额外的GS接头和二分NLS序列);3.密码子优化:对TnpBmax系统进行了哺乳动物密码子优化 | 利用TEEP将原始的ωRNA支架缩短至117个核苷酸,在ωRNA的3′端融合HDV核酶得到mini-ωRNA | TnpBmax系统的RuvC结构域中引入了突变(D191A)并融合了TadA8e16生成碱基编辑器 | 编辑率比野生型提高了1.3倍;在小鼠肝脏中实现了高达75.3%的编辑效率,在小鼠脑中实现了高达65.9%的编辑效率 | 编辑效率可达16.6% | [ |
| 2025/2/16 | ISDra2 TnpB系统 | 408 | 231 | HEK293T;本氏烟草 | 1.氨基酸替换:P282I单点突变使效率提升四倍;深度突变扫描(DMS)筛选到6个组合突变的变体(eTnpB1a-eTnpB1f),eTnpB1e效率最高;2.蛋白结构域截短:C端截短至376 aa,是最小活性TnpB截断变体 | 茎2区域的“铰链”区域(rA-37-rU-44)发生的碱基缺失和替换编辑效率最高 | 无 | eTnpB1e在PDS1-1位点编辑效率为33%,与WT ISDra2 TnpB (< 1%)相比增加了50倍以上 | 无 | [ |
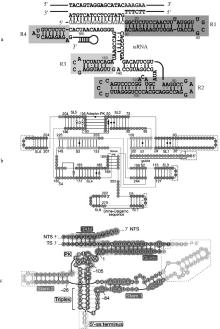
Fig. 4
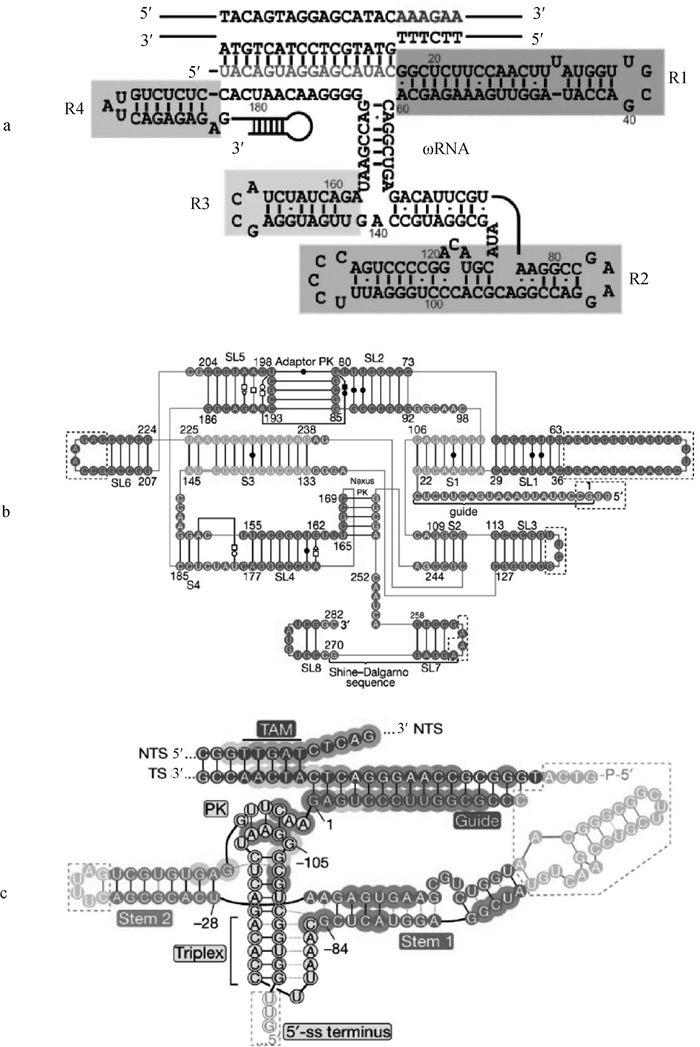
Predicted secondary structures of the guide RNAs from the IscB, IsrB and TnpB systems [23, 32, 36] a. The ωRNA of the IscB system is divided into 4 regions (R1, R2, R3, and R4);b. The ωRNA of the IsrB system is divided into 4 stems (S1-4) and 8 stem-loops (SL1-8); the adaptor PK is the junction pseudoknot structure formed by SL2 and SL5; the nexus PK is the nexus pseudoknot structure formed by the region between SL4, S2, and SL7; the Shine-Dalgarno sequence is a ribosome binding site in bacteria, located approximately 8 bases upstream of the mRNA start codon (usually AUG); c. The reRNA of the TnpB system is divided into stem 1 and 2; Triplex is triple helix structure; Stem is stem-loop structure; 5′-ss terminus is 5′ end terminal sequence; PK is pseudoknot structure; TS is target strand; NTS is non-target strand; TAM is target-adjacent motif"
| 1 |
GALLAGHERD N,HABERJ E.Repair of a site-specific DNA cleavage: Old-school lessons for Cas9-mediated gene editing[J].ACS Chem Biol,2018,13(2):397-405.
doi: 10.1021/acschembio.7b00760 |
| 2 |
SCHERERS,DAVISR W.Replacement of chromosome segments with altered DNA sequences constructed in vitro[J].Proc Natl Acad Sci U S A,1979,76(10):4951-4955.
doi: 10.1073/pnas.76.10.4951 |
| 3 |
DOYONY,VOT D,MENDELM C,et al.Enhancing zinc-finger-nuclease activity with improved obligate heterodimeric architectures[J].Nat Methods,2011,8(1):74-79.
doi: 10.1038/nmeth.1539 |
| 4 |
王玮玮,刘瑞琪,吴勇延,等.CRISPR/Cas9基因编辑系统研究进展及其在动物基因编辑研究中的应用[J].畜牧兽医学报,2016,47(7):1299-1305.
doi: 10.11843/j.issn.0366-6964.2016.07.001 |
|
WANGW W,LIUR Q,WUY Y,et al.The Progress of CRISPR/Cas9 System and Its Application in Animal Genetic Engineering[J].Acta Veterinaria et Zootechnica Sinica,2016,47(7):1299-1305.
doi: 10.11843/j.issn.0366-6964.2016.07.001 |
|
| 5 |
CUIX,JID,FISHERD A,et al.Targeted integration in rat and mouse embryos with zinc-finger nucleases[J]. Nat Biotechnol,2011,29(1):64-67.
doi: 10.1038/nbt.1731 |
| 6 |
MILLERJ C,TANS,QIAOG,et al.A TALE nuclease architecture for efficient genome editing[J].Nat Biotechnol,2011,29(2):143-148.
doi: 10.1038/nbt.1755 |
| 7 |
CERMAKT,DOYLEE L,CHRISTIANM,et al.Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting[J].Nucleic Acids Res,2011,39(12):e82.
doi: 10.1093/nar/gkr218 |
| 8 |
HOCKEMEYERD,WANGH,KIANIS,et al.Genetic engineering of human pluripotent cells using TALE nucleases[J].Nat Biotechnol,2011,29(8):731-734.
doi: 10.1038/nbt.1927 |
| 9 | ZHANGF,VOYTASD F.Targeted mutagenesis in Arabidopsis using zinc-finger nucleases[J].Methods Mol Biol,2011,701,167-177. |
| 10 |
WHYTEJ J,ZHAOJ,WELLSK D,et al.Gene targeting with zinc finger nucleases to produce cloned eGFP knockout pigs[J].Mol Reprod Dev,2011,78(1):2.
doi: 10.1002/mrd.21271 |
| 11 |
WIEDENHEFTB,STERNBERGS H,DOUDNAJ A.RNA-guided genetic silencing systems in bacteria and archaea[J]. Nature,2012,482(7385):331-338.
doi: 10.1038/nature10886 |
| 12 |
CONGL,RANF A,COXD,et al.Multiplex genome engineering using CRISPR/Cas systems[J].Science,2013,339(6121):819-823.
doi: 10.1126/science.1231143 |
| 13 |
ALTAE-TRANH,KANNANS,DEMIRCIOGLUF E,et al.The widespread IS200/IS605 transposon family encodes diverse programmable RNA-guided endonucleases[J].Science,2021,374(6563):57-65.
doi: 10.1126/science.abj6856 |
| 14 |
KANNANS,ALTAE-TRANH,ZHUS,et al.Evolution-guided protein design of IscB for persistent epigenome editing in vivo[J].Nat Biotechnol,2025,
doi: 10.1038/s41587-025-02655-3 |
| 15 |
WANGF,MAS,ZHANGS,et al.CRISPR beyond: harnessing compact RNA-guided endonucleases for enhanced genome editing[J].Sci China Life Sci,2024,67(12):2563-2574.
doi: 10.1007/s11427-023-2566-8 |
| 16 |
KONGX,LIT,YANGH.AAV-mediated gene therapies by miniature gene editing tools[J].Sci China Life Sci,2024,67(12):2540-2553.
doi: 10.1007/s11427-023-2608-5 |
| 17 |
KATOK,OKAZAKIS,KANNANS,et al.Structure of the IscB-ωRNA ribonucleoprotein complex, the likely ancestor of CRISPR-Cas9[J]. Nat Commun,2022,13(1):6719.
doi: 10.1038/s41467-022-34378-3 |
| 18 | XUC,NIUX,SUNH,et al.Conversion of IscB and Cas9 into RNA-guided RNA editors[J].Cell,2025,S0092-8674(25):00854-2. |
| 19 | KAPITONOVV V,MAKAROVAK S,KOONINE V.ISC, a novel group of bacterial and archaeal DNA transposons that encode Cas9 homologs[J].J Bacteriol,2015,198(5):797-807. |
| 20 |
WANGK,WANGJ,YANGX,et al.Structural insights into Type Ⅱ-D Cas9 and its robust cleavage activity[J].Nat Commun,2025,16(1):7396.
doi: 10.1038/s41467-025-62128-8 |
| 21 |
GAOJ,WANGH,SUNJ,et al.Highly efficient genome editing in Bacillus subtilis via miniature DNA nucleases IscB[J]. Synth Syst Biotechnol,2025,10(4):1215-1223.
doi: 10.1016/j.synbio.2025.06.012 |
| 22 |
LVJ,JINJ,DINGL,et al.Directed evolution of ogeuiscb with enhanced activity in human cells[J].FASEB J,2025,39(8):e70570.
doi: 10.1096/fj.202500082R |
| 23 |
HIRANOS,KAPPELK,ALTAE-TRANH,et al.Structure of the OMEGA nickase IsrB in complex with ωRNA and target DNA[J].Nature,2022,610(7932):575-581.
doi: 10.1038/s41586-022-05324-6 |
| 24 |
YUZ,SHEQ.Genome editing from Cas9 to IscB: Backwards and forwards towards new breakthroughs[J].Eng Microbiol,2021,1,100004.
doi: 10.1016/j.engmic.2021.100004 |
| 25 |
MARQUARTK F,MATHISN,MOLLAYSAA,et al.Effective genome editing with an enhanced ISDra2 TnpB system and deep learning-predicted ωRNAs[J].Nat Methods,2024,21(11):2084-2093.
doi: 10.1038/s41592-024-02418-z |
| 26 |
JIANGK,LIMJ,SGRIZZIS,et al.Programmable RNA-guided endonucleases are widespread in eukaryotes and their viruses[J]. bioRxiv[Preprint],2023,
doi: 10.1101/2023.06.13.544871 |
| 27 |
KARVELIST,DRUTEIKAG,BIGELYTEG,et al.Transposon-associated TnpB is a programmable RNA-guided DNA endonuclease[J].Nature,2021,599(7886):692-696.
doi: 10.1038/s41586-021-04058-1 |
| 28 |
FUW,MAJ,WANGZ,et al.Mechanisms and engineering of a miniature type V-N CRISPR-Cas12 effector enzyme[J].Nat Commun,2025,16(1):5667.
doi: 10.1038/s41467-025-61290-3 |
| 29 |
SAJJADM W,NAQVIR Z,AMINI.Plant genome editing goes viral[J].Trends Biotechnol,2025,43(7):1520-1522.
doi: 10.1016/j.tibtech.2025.05.005 |
| 30 |
WEIY,GAOP,PAND,et al.Engineering eukaryotic transposon-encoded Fanzor2 system for genome editing in mammals[J]. Nat Chem Biol,2025,
doi: 10.1038/s41589-025-01902-7 |
| 31 |
JIANGK,GOOTENBERGJ S,ABUDAYYEHO O.Fanzors, a family of eukaryotic RNA-guided DNA endonucleases[J].FEBS Lett,2025,599(8):1089-1093.
doi: 10.1002/1873-3468.70038 |
| 32 |
SASNAUSKASG,TAMULAITIENEG,DRUTEIKAG,et al.TnpB structure reveals minimal functional core of Cas12 nuclease family[J].Nature,2023,616(7956):384-389.
doi: 10.1038/s41586-023-05826-x |
| 33 | KLEINSTIVERB P,SOUSAA A,WALTONR T,et al.Engineered CRISPR-Cas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing[J].Nat Biotechnol,2020,38(7):901. |
| 34 | ZHANGH,KONGX,XUEM,et al.An engineered xCas12i with high activity, high specificity, and broad PAM range[J].Protein Cell,2023,14(7):538-543. |
| 35 |
CHATTERJEEP,JAKIMON,LEEJ,et al.An engineered ScCas9 with broad PAM range and high specificity and activity[J].Nat Biotechnol,2020,38(10):1154-1158.
doi: 10.1038/s41587-020-0517-0 |
| 36 |
HAND,XIAOQ,WANGY,et al.Development of miniature base editors using engineered IscB nickase[J].Nat Methods,2023,20(7):1029-1036.
doi: 10.1038/s41592-023-01898-9 |
| 37 |
XUEN,HONGD,ZHANGD,et al.Engineering IscB to develop highly efficient miniature editing tools in mammalian cells and embryos[J]. Mol Cell,2024,84(16):3128-3140.
doi: 10.1016/j.molcel.2024.07.007 |
| 38 |
STOTTK,WATSONM,BOSTOCKM J,et al.Structural insights into the mechanism of negative regulation of single-box high mobility group proteins by the acidic tail domain[J]. J Biol Chem,2014,289(43):29817-29826.
doi: 10.1074/jbc.M114.591115 |
| 39 |
KLASSJ,MURPHYFV 4TH,FOUTSS,et al.The role of intercalating residues in chromosomal high-mobility-group protein DNA binding, bending and specificity[J].Nucleic Acids Res,2003,31(11):2852-2864.
doi: 10.1093/nar/gkg389 |
| 40 |
YINS,ZHANGM,LIUY,et al.Engineering of efficiency-enhanced Cas9 and base editors with improved gene therapy efficacies[J].Mol Ther,2023,31(3):744-759.
doi: 10.1016/j.ymthe.2022.11.014 |
| 41 |
YANH,TANX,ZOUS,et al.Assessing and engineering the IscB-ωRNA system for programmed genome editing[J].Nat Chem Biol,2024,20(12):1617-1628.
doi: 10.1038/s41589-024-01669-3 |
| 42 |
XIAOQ,LIG,HAND,et al.Engineered IscB-ωRNA system with expanded target range for base editing[J].Nat Chem Biol,2025,21(1):100-108.
doi: 10.1038/s41589-024-01706-1 |
| 43 | THORNTON B W, WEISSMAN R F, TRAN R V, et al. Latent activity in TnpB revealed by mutational scanning[J]. bioRxiv[Preprint], 2025, 637750. |
| 44 |
KIMD Y,CHUNGY,LEEY,et al.Hypercompact adenine base editors based on transposase B guided by engineered RNA[J].Nat Chem Biol,2022,18(9):1005-1013.
doi: 10.1038/s41589-022-01077-5 |
| 45 |
CHENW,MAJ,WUZ,et al.Cas12n nucleases, early evolutionary intermediates of type V CRISPR, comprise a distinct family of miniature genome editors[J].Mol Cell,2023,83(15):2768-2780.
doi: 10.1016/j.molcel.2023.06.014 |
| 46 |
ALTAE-TRANH,SHMAKOVS A,MAKAROVAK S,et al.Diversity, evolution, and classification of the RNA-guided nucleases TnpB and Cas12[J].Proc Natl Acad Sci U S A,2023,120(48):e2308224120.
doi: 10.1073/pnas.2308224120 |
| 47 |
WANGM,SUNZ,LIUY,et al.Hypercompact TnpB and truncated TnpB systems enable efficient genome editing in vitro and in vivo[J].Cell Discov,2024,10(1):31.
doi: 10.1038/s41421-023-00645-w |
| 48 |
WUY,YUANQ,ZHUY,et al.Improving FnCas12a genome editing by exonuclease fusion[J].CRISPR J,2020,3(6):503-511.
doi: 10.1089/crispr.2020.0073 |
| 49 |
YINJ,LUR,XINC,et al.Cas9 exo-endonuclease eliminates chromosomal translocations during genome editing[J].Nat Commun,2022,13(1):1204.
doi: 10.1038/s41467-022-28900-w |
| 50 |
WANGW,LIS,YANGJ,et al.Exploiting the efficient Exo: Cas12i3-5M fusions for robust single and multiplex gene editing in rice[J].J Integr Plant Biol,2025,67(5):1246-1253.
doi: 10.1111/jipb.13850 |
| 51 |
WANGW,YANL,LIJ,et al.Engineering a robust Cas12i3 variant-mediated wheat genome editing system[J].Plant Biotechnol J,2025,23(3):860-873.
doi: 10.1111/pbi.14544 |
| 52 |
LIAOK,CHENK,MAS,et al.exoCasMINI: A T5 exonuclease fused CRISPR-Cas12f system with enhanced gene editing efficiency[J].iScience,2025,28(8):113171.
doi: 10.1016/j.isci.2025.113171 |
| 53 |
MAS,LIAOK,CHENK,et al.hpCasMINI: An engineered hypercompact CRISPR-Cas12f system with boosted gene editing activity[J].Nat Commun,2025,16(1):5001.
doi: 10.1038/s41467-025-60124-6 |
| 54 | REZAEI S, MONCADA-RESTREPO M, LENG S, et al. Synthesizing unmodified, supercoiled circular DNA molecules in vitro[J]. bioRxiv[Preprint], 2025, 01.24.634800. |
| 55 |
SHUIS,WANGS,LIUJ.Systematic investigation of the effects of multiple SV40 nuclear localization signal fusion on the genome editing activity of purified SpCas9[J]. Bioengineering (Basel),2022,9(2):83.
doi: 10.3390/bioengineering9020083 |
| 56 |
NⅡNUMAS,WAKEY,NAKAGAWAY,et al.Importance of nuclear localization signal-fused Cas9 in the production of genome-edited mice via embryo electroporation[J].Biochem Biophys Res Commun,2023,685,149140.
doi: 10.1016/j.bbrc.2023.149140 |
| 57 |
SALIMI-JEDAA,ESGHAEIM,HOSSEINKEYVANI,et al.Inhibition of HIV-1 replication using the CRISPR/cas9-no NLS system as a prophylactic strategy[J].Heliyon,2022,8(9):e10483.
doi: 10.1016/j.heliyon.2022.e10483 |
| 58 |
LVZ,CHENW,FANGS,et al.Targeted mutagenesis in Arabidopsis and medicinal plants using transposon-associated TnpB[J].J Integr Plant Biol,2024,66(10):2083-2086.
doi: 10.1111/jipb.13758 |
| 59 |
WANGL,HANH.Strategies for improving the genome-editing efficiency of class 2 CRISPR/Cas system[J].Heliyon,2024,10(19):e38588.
doi: 10.1016/j.heliyon.2024.e38588 |
| 60 |
NALLEYM J,BANERJEES,HUANGM Y,et al.Near 100% efficient homology-dependent genome engineering in the human fungal pathogen Cryptococcus neoformans[J].G3 (Bethesda),2025,15(8):jkaf118.
doi: 10.1093/g3journal/jkaf118 |
| 61 |
WALLACEK A,GERSTENBERGT L,ENNISC L,et al.A differentiated β-globin gene replacement strategy uses heterologous introns to restore physiological expression[J].Mol Ther,2025,33(4):1407-1419.
doi: 10.1016/j.ymthe.2025.02.036 |
| 62 |
KARMAKARS,PANDAD,PANDAS,et al.A miniature alternative to Cas9 and Cas12: Transposon-associated TnpB mediates targeted genome editing in plants[J].Plant Biotechnol J,2024,22(10):2950-2953.
doi: 10.1111/pbi.14416 |
| 63 |
ZHANGY,RENQ,TANGX,et al.Expanding the scope of plant genome engineering with Cas12a orthologs and highly multiplexable editing systems[J].Nat Commun,2021,12(1):1944.
doi: 10.1038/s41467-021-22330-w |
| 64 |
ZHAOY,BOEKEJ D.CRISPR-Cas12a system in fission yeast for multiplex genomic editing and CRISPR interference[J].Nucleic Acids Res,2020,48(10):5788-5798.
doi: 10.1093/nar/gkaa329 |
| 65 |
ZHONGZ,LIUS,LIUX,et al.Intron-based single transcript unit CRISPR systems for plant genome editing[J].Rice (N Y),2020,13(1):8.
doi: 10.1186/s12284-020-0369-8 |
| 66 |
ZHANGR,TANGX,HEY,et al.IsDge10 is a hypercompact TnpB nuclease that confers efficient genome editing in rice[J].Plant Commun,2024,5(11):101068.
doi: 10.1016/j.xplc.2024.101068 |
| 67 |
JINEKM,CHYLINSKIK,FONFARAI,et al.A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity[J].Science,2012,337(6096):816-821.
doi: 10.1126/science.1225829 |
| 68 |
GUOR,SUNX,WANGF,et al.Engineered IscB-ωRNA system with improved base editing efficiency for disease correction via single AAV delivery in mice[J].Cell Rep,2024,43(11):114973.
doi: 10.1016/j.celrep.2024.114973 |
| 69 |
HANL,HUY,MOQ,et al.Engineering miniature IscB nickase for robust base editing with broad targeting range[J].Nat Chem Biol,2024,20(12):1629-1639.
doi: 10.1038/s41589-024-01670-w |
| 70 |
LIZ,GUOR,SUNX,et al.Engineering a transposon-associated TnpB-ωRNA system for efficient gene editing and phenotypic correction of a tyrosinaemia mouse model[J]. Nat Commun,2024,15(1):831.
doi: 10.1038/s41467-024-45197-z |
| 71 |
RICHTERM F,ZHAOK T,ETONE,et al.Phage-assisted evolution of an adenine base editor with improved Cas domain compatibility and activity[J].Nat Biotechnol,2020,38(7):883-891.
doi: 10.1038/s41587-020-0453-z |
| 72 |
ARANTESP R,CHENX,SINHAS,et al.Dimerization of the deaminase domain and locking interactions with Cas9 boost base editing efficiency in ABE8e[J].Nucleic Acids Res,2024,52(22):13931-13944.
doi: 10.1093/nar/gkae1066 |
| 73 |
包斌武,邹惠影,李俊良,等.基因编辑技术的研究进展[J].畜牧兽医学报,2025,56(1):1-14.
doi: 10.11843/j.issn.0366-6964.2025.01.001 |
|
BAOB W,ZOUH Y,LIJ L,et al.Research progress in gene editing technology[J].Acta Veterinaria et Zootechnica Sinica,2025,56(1):1-14.
doi: 10.11843/j.issn.0366-6964.2025.01.001 |
|
| 74 |
WANGY,PROSENDE,MEIL,et al.A novel strategy to engineer DNA polymerases for enhanced processivity and improved performance in vitro[J].Nucleic Acids Res,2004,32(3):1197-1207.
doi: 10.1093/nar/gkh271 |
| [1] | SHAO Jiahao, ZHANG Yanjie, ZHAO Yongju. Research Progress of N6-methyladenosine (m6A) Modification in Regulation of Livestock and Poultry Genetic Breeding and Reproduction [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(10): 4774-4786. |
| [2] | CAO Yu, ZHOU Bohan, XU Qi, YUAN Zi'ao, SU Rui, LÜ Qi, LI Jinquan, ZHANG Yanjun, WANG Ruijun, WANG Zhiying. Research Progress on Integrated eQTL-GWAS Data Analysis for Potential Functional Genetic Loci Identification in Animal Breeding [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(10): 4759-4773. |
| [3] | QIAN Li, LIANG Mang, DENG Tianyu, DU Lili, LI Keanning, QIU Shiyuan, XUE Qingqing, ZHANG Lupei, GAO Xue, XU Lingyang, ZHENG Caihong, LI Junya, GAO Huijiang. Improving Genomic Prediction Accuracy via Auto-encoder-based Compression of Transcriptome Data [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(9): 4410-4421. |
| [4] | LI Jiapeng, LIU Qing, SUN Jiayu, MA Zefang, CUI Kai. Screening of Key Genes for Coat Color Formation in Silver Fox Based on Transcriptome and Proteome Analyses [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(9): 4379-4392. |
| [5] | ZHENG Yunchang, HOU Ruilin, LIANG Xiaohe, YANG Lidan, ZHANG Yinjiao, HUO Haonan, CHEN Weina, ZHANG Cui, LI Shijie. Analysis of Monoallelic Expression and DNA Methylation Status of Bovine FOXP2 Gene [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(9): 4369-4378. |
| [6] | WANG Yanbo, ZHANG Xiaomeng, JING Xiujuan, FENG Xiaoyi, ZHANG Yuanqing, ZHAO Xueming. Advances in Nanoparticle Applications for Animal Germplasm Cryopreservation [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(9): 4156-4164. |
| [7] | ZHANG Yang, WANG Zhongfa, LI Minjuan, HE Yunan, GUAN Weijun. Cultivation and Identification of Tenogenic Mesenchymal Stem Cells for Sports-Related Injury Therapy in vitro [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(8): 3813-3825. |
| [8] | ZHANG Fan, ZENG Wei, ZHOU Ao. Advances in Gene Editing for Disease Resistance Breeding in Livestock and Poultry [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(7): 3047-3056. |
| [9] | ZHOU Rui, WU De, CHE Lianqiang, LIN Yan, FENG Bin, FANG Zhengfeng. Advances of N6-Adenosine Methylation Regulating Adipogenesis [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(5): 1995-2003. |
| [10] | BAO Binwu, ZOU Huiying, LI Junliang, GAO Chen, GAO Huijiang, DU Zhenwei, ZHANG Boyu, LI Junya, GAO Xue. Research Progress in Gene Editing Technology [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(1): 1-14. |
| [11] | MIN Xiangyu, WEI Jiali, XU Biao, LIU Huitao, ZHENG Junjun, WANG Guiwu. Full-length Transcriptome Sequencing of Sika Deer Antler and Mining of Antler Yield-related Genes [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(12): 5549-5566. |
| [12] | Hongxia JIA, Zaixia LIU, Le ZHOU, Yanchun BAO, Chenxi HUO, Pengpeng ZUO, Mingjuan GU, Risu NA, Wenguang ZHANG. Research Progress of Genomic Selection in Beef Cattle [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(9): 3757-3768. |
| [13] | Zhentao XIA, Nan WANG, Wanjie WANG, Qilü ZHOU, Lei HUANG, Yulian MU. Characteristics Analysis of TGEV Infection Mediated by IPEC-J2 with Knockout of pAPN Gene [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(8): 3395-3407. |
| [14] | Xiaoxu ZHANG, Hao LI, Pingjie FENG, Hao YANG, Xinyue LI, Ran LÜ, Zhangyuan PAN, Mingxing CHU. Application of Single-Cell Transcriptome Sequencing Technology in Domesticated Animals [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(8): 3276-3287. |
| [15] | Wenwen LIU, Faming DONG, Yanzhen BI. The Development of Multi-Gene Editing Technology and Its Application in Agricultural Biological Germplasm Innovation [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(8): 3267-3275. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||