





Acta Veterinaria et Zootechnica Sinica ›› 2025, Vol. 56 ›› Issue (9): 4604-4614.doi: 10.11843/j.issn.0366-6964.2025.09.038
• Preventive Veterinary Medicine • Previous Articles Next Articles
YU Qiurong1,2( ), CAI Xuhang2, HE Yi3, LI Jizong2, MAO Li2,*(
), CAI Xuhang2, HE Yi3, LI Jizong2, MAO Li2,*( ), XU Xingang1,*(
), XU Xingang1,*( ), LI Bin2
), LI Bin2
Received:2024-10-14
Online:2025-09-23
Published:2025-09-30
Contact:
MAO Li, XU Xingang
E-mail:yuqiurong2233@163.com;mao-li@live.cn;tiger2003@nwsuaf.edn.cn
CLC Number:
YU Qiurong, CAI Xuhang, HE Yi, LI Jizong, MAO Li, XU Xingang, LI Bin. Identification and Isolation of a Caprine Coronavirus and Analysis of the Complete Genome Sequence[J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(9): 4604-4614.
Table 1
Primers sequences for the S and NS4a-4b gene amplification"
| 引物名称 Primer name | 引物序列(5′→3′) Primer sequence | 扩增产物大小/bp Product size |
| cpCoV-S1-F | TGCTGTTATAGGAGATTTAAAGT | 1 403 |
| cpCoV-S1-R | GCTTTAAAACAATGTTGTGCATA | |
| cpCoV-S2-F | GCTGCTAATGTTTCTGTTAGC | 1 517 |
| cpCoV-S2-R | TCCAGTGCAATTATTATAAGCC | |
| cpCoV-S3-F | GATCTGCTATAGAGGATTTAC | 1 330 |
| cpCoV-S3-R | TAATTACTAACTCCTGGTGTC | |
| cpCoV-NS4ab-F | TCTTCATATGCTGTTGTACAGG | 1 038 |
| cpCoV-NS4ab-R | TCCACATCAAGAACTGGTGGT |

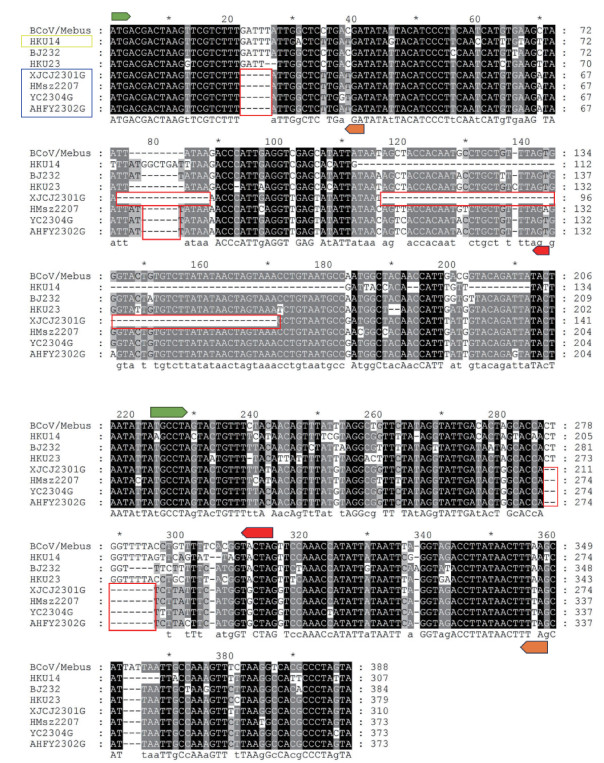
Fig. 6
Alignment results of NS4a-4b sequences of cpCoV and reference strains Green logo indicates the start codons; Orange logo indicates the stop codons for cpCoV/XJCJ2301G; Red logo indicates the stop codons for BCoV/Mebus; Red box indicates the base deletion compared to BCoV/Mebus; Blue box indicates the cpCoV reference strains; Yellow box indicates the reference strain of rabbit coronavirus"
| 1 |
THOMASC J,HOETA E,SREEVATSANS,et al.Transmission of bovine coronavirus and serologic responses in feedlot calves under field conditions[J].Am J Vet Res,2006,67(8):1412-1420.
doi: 10.2460/ajvr.67.8.1412 |
| 2 | VLASOVAA N,SAIFL J.Bovine coronavirus and the associated diseases[J].Front Vet Sci,2021,31(8):643220. |
| 3 |
CEBRAC K,MATTSOND E,BAKERR J,et al.Potential pathogens in feces from unweaned llamas and alpacas with diarrhea[J].J Am Vet Med Assoc,2003,223(12):1806-1808.
doi: 10.2460/javma.2003.223.1806 |
| 4 |
CHASEYD,REYNOLDSD J,BRIDGERJ C,et al.Identification of coronaviruses in exotic species of Bovidae[J].Vet Rec,1984,115(23):602-603.
doi: 10.1136/vr.115.23.602 |
| 5 |
HASOKSUZM,ALEKSEEVK,VLASOVAA,et al.Biologic, antigenic, and full-length genomic characterization of a bovine-like coronavirus isolated from a giraffe[J].J Virol,2007,81(10):4981-4990.
doi: 10.1128/JVI.02361-06 |
| 6 |
TSUNEMITSUH,EL-KANAWATIZ R,SMITHD R,et al.Isolation of coronaviruses antigenically indistinguishable from bovine coronavirus from wild ruminants with diarrhea[J].J Clin Microbiol,1995,33(12):3264-3269.
doi: 10.1128/jcm.33.12.3264-3269.1995 |
| 7 |
ALEKSEEVK,VLASOVAA,JUNGK,et al.Bovine-like coronaviruses isolated from four species of captive wild ruminants are homologous to bovine coronaviruses, based on complete genomic sequences[J].J Virol,2008,82(24):12422-12431.
doi: 10.1128/JVI.01586-08 |
| 8 |
CLARKM.Bovine coronavirus[J].Br Vet J,1993,149(1):51-70.
doi: 10.1016/S0007-1935(05)80210-6 |
| 9 |
HODNIKJ,JEŽEKJ,STARI AČGJ.Coronaviruses in cattle[J].Trop Anim Health Prod,2020,52(6):2809-2816.
doi: 10.1007/s11250-020-02354-y |
| 10 | 黄金,李思远,谢玲玲,等.牛冠状病毒TaqMan荧光定量检测方法的建立及国内部分地区流行病学调查[J].江苏农业科学,2023,51(17):29-33. |
| HUANGJ,LIS Y,XIEL L,et al.Establishment of TaqMan fluorescent quantitative detection method for bovine coronavirus and epidemiological investigation in some regions of China[J].Jiangsu Agricultural Sciences,2023,51(17):29-33. | |
| 11 |
ZHUQ,LIB,SUND.Advances in bovine coronavirus epidemiology[J].Viruses,2022,14(5):1109.
doi: 10.3390/v14051109 |
| 12 | 姚火春,杜念兴,徐为燕,等.牛冠状病毒的血清流行病学调查[J].南京农业大学学报,1990(2):117-121. |
| YAOH C,DUN X,XUW Y,et al.Serological epidemiological survey of bovine coronavirus[J].Journal of Nanjing Agricultural University,1990(2):117-121. | |
| 13 |
LOJKI AC'U1I,KREŠI AC'U1N,ŠIMI AC'U1I,et al.Detection and molecular characterisation of bovine corona and toroviruses from Croatian cattle[J].BMC Vet Res,2015,11,202.
doi: 10.1186/s12917-015-0511-9 |
| 14 |
UHDEF L,KAUFMANNT,SAGERH,et al.Prevalence of four enteropathogens in the faeces of young diarrhoeic dairy calves in Switzerland[J].Vet Rec,2008,163(12):362-366.
doi: 10.1136/vr.163.12.362 |
| 15 |
IZZOM M,KIRKLANDP D,MOHLERV L,et al.Prevalence of major enteric pathogens in Australian dairy calves with diarrhoea[J].Aust Vet J,2011,89(5):167-173.
doi: 10.1111/j.1751-0813.2011.00692.x |
| 16 |
REDDAY T,ADAMUH,BERGHOLMJ,et al.Detection and characterization of bovine coronavirus and rotavirus in calves in Ethiopia[J].BMC Vet Res,2025,21(1):122.
doi: 10.1186/s12917-025-04563-9 |
| 17 | BIDOKHTIM,TRÅVÉNM,KRISHNAN,et al.Evolutionary dynamics of bovine coronaviruses: natural selection pattern of the spike gene implies adaptive evolution of the strains[J].J Gen Virol,2013,94(Pt 9):2036-2049. |
| 18 |
ZHAOL,WANGD,JIANGH,et al.Isolation and characterization of bovine coronavirus variants with mutations in the hemagglutinin-esterase gene in dairy calves in China[J].BMC Vet Res,2025,21(1):92.
doi: 10.1186/s12917-025-04538-w |
| 19 |
ZHANGF,CHAIC,NIUR,et al.Genetic characterization of bovine coronavirus strain isolated in Inner Mongolia of China[J].BMC Vet Res,2024,20(1):209.
doi: 10.1186/s12917-024-04046-3 |
| 20 |
SHAHA U,GAUGERP,HEMIDAM G.Isolation and molecular characterization of an enteric isolate of the genotype-Ia bovine coronavirus with notable mutations in the receptor binding domain of the spike glycoprotein[J].Virology,2025,603,110313.
doi: 10.1016/j.virol.2024.110313 |
| 21 | 宋广林,董惠兰,滕庆,等.犊牛流行性腹泻病原研究[J].畜牧兽医学报,1985,16(2):121-124. |
| SONGG L,DONGH L,TENGQ,et al.Pathogen research on calf diarrhea disease[J].Acta Veterinaria et Zootechnica Sinica,1985,16(2):121-124. | |
| 22 |
黄金,李思远,毛立,等.牛冠状病毒S1蛋白的真核表达及间接ELISA方法的建立与应用[J].畜牧兽医学报,2024,55(5):2050-2060.
doi: 10.11843/j.issn.0366-6964.2024.05.023 |
|
HUANGJ,LIS Y,MAOL,et al.Eukaryotic expression of bovine coronavirus S1 protein and establishment and application of indirect ELISA[J].Acta Veterinaria et Zootechnica Sinica,2024,55(5):2050-2060.
doi: 10.11843/j.issn.0366-6964.2024.05.023 |
|
| 23 |
LIUQ,NIUX,JIANGL,et al.Establishment of an indirect ELISA method for detecting bovine coronavirus antibodies based on N protein[J].Front Vet Sci,2025,12,1530870.
doi: 10.3389/fvets.2025.1530870 |
| 24 |
SUZUKIT,OTAKEY,UCHIMOTOS,et al.Genomic characterization and phylogenetic classification of bovine coronaviruses through whole genome sequence analysis[J].Viruses,2020,12(2):183.
doi: 10.3390/v12020183 |
| 25 |
LIF.Structure, function, and evolution of coronavirus spike proteins[J].Annu Rev Virol,2016,3(1):237-261.
doi: 10.1146/annurev-virology-110615-042301 |
| 26 |
BIDOKHTIM,BAULEC,HAKHVERDYANM,et al.Tracing the transmission of bovine coronavirus infections in cattle herds based on S gene diversity[J].Vet J,2012,193(2):386-390.
doi: 10.1016/j.tvjl.2011.12.015 |
| 27 |
DECARON,CAMPOLOM,DESARIOC,et al.Respiratory disease associated with bovine coronavirus infection in cattle herds in Southern Italy[J].J Vet Diagn Invest,2008,20(1):28-32.
doi: 10.1177/104063870802000105 |
| 28 |
VIJGENL,KEYAERTSE,LEMEYP,et al.Circulation of genetically distinct contemporary human coronavirus OC43 strains[J].Virology,2005,337(1):85-92.
doi: 10.1016/j.virol.2005.04.010 |
| 29 |
MILLETJ K,JAIMESJ A,WHITTAKERG R.Molecular diversity of coronavirus host cell entry receptors[J].FEMS Microbiol Rev,2021,45(3):fuaa057.
doi: 10.1093/femsre/fuaa057 |
| 30 |
KLEINE-WEBERH,ELZAYATM,HOFFMANNM,et al.Functional analysis of potential cleavage sites in the MERS-coronavirus spike protein[J].Sci Rep,2018,8(1):16597.
doi: 10.1038/s41598-018-34859-w |
| 31 |
TANGT,JAIMESJ,BIDONM,et al.Proteolytic activation of SARS-CoV-2 spike at the S1/S2 boundary: potential role of proteases beyond furin[J].ACS Infect Dis,2021,7(2):264-272.
doi: 10.1021/acsinfecdis.0c00701 |
| 32 |
ABRAHAMS,KIENZLET,LAPPSW,et al.Sequence and expression analysis of potential nonstructural proteins of 4.9, 4.8, 12.7, and 9.5 kDa encoded between the spike and membrane protein genes of the bovine coronavirus[J].Virology,1990,177(2):488-495.
doi: 10.1016/0042-6822(90)90513-Q |
| [1] | CHEN Yunlong, FAN Gang, FAN Xinyi, GUO Yongchao, ZHANG Xinmiao, WANG Yan, ZHANG Shiqiang. Isolation and Identification of a Novel Porcine Circovirus Type 2d Strain with Multiple Nucleotide Substitutions [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(6): 3032-3040. |
| [2] | WANG Yanan, GUO Yaru, JIANG Yanping, CUI Wen, LI Jiaxuan, LI Yijing, WANG Li. Isolation, Identification and Pathogenicity Analysis of Porcine Rotavirus [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(5): 2259-2269. |
| [3] | WANG Jinxiang, SU Jinbo, FU Huanru, SUN Shikun, GAO Chengfang, CHEN Dongjin, SANG Lei, XIE Xiping. Pathogenicity and Genomic Features of Rabbit Sourced Serogroup A Pasteurella multocida Isolates Pm3 and Pm6 [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(5): 2340-2352. |
| [4] | KUANG Yinzhi, XU Fengpei, ZHOU Pei. Whole Genome Sequence Analysis of Canine Circovirus in an Otter (Lutra lutra) [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(4): 1989-1994. |
| [5] | LIU Jian, YU Zehai, ZHANG Meiyu, LI Dan, WANG Jun, LIU Fangqin, ZHANG Qun, XU Shouzhen. Full-genome Analysis of a Bovine Enterovirus Type F and the Establishment of an Indirect ELISA Method for Antibody Detection [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(2): 814-825. |
| [6] | DU Qingjie, WU Liping, ZHANG Fan, DAI Pengxiu, FENG Xiancheng, ZHANG Xinke. Difference Analysis of Oral Flora in Dogs with Periodontitis and Drug Resistance of Oral Porphyromonas [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(2): 934-942. |
| [7] | WU Pingxian, WANG Junge, DIAO Shuqi, CHAI Jie, ZHA Lin, GUO Zongyi, CHEN Hongyue, LONG Xi. Analysis of Genetic Architecture Characteristics and Selection Signature by Imputed Whole Genome Sequencing Data in Rongchang Pigs [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(1): 147-158. |
| [8] | Shan ZHANG, Dahu LIU, Baojing LIU, Lin LIANG, Ruiying LIANG, Xinming TANG, Xusheng QIU, Chan DING, Jiabo DING, Shaohua HOU. Isolation, Identification and Pathogenicity Analysis of a Pigeon Paramyxovirus-1 Strain [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(9): 4051-4060. |
| [9] | Cheng YANG, Ye LIU, Ning CHENG, Kaiyue WANG, Xinlei LI, Jiuying SUN, Junping HAN, Wenjun LI, Huanhuan WANG, Xiao SHAO, Xuejiao CHENG, Yingfeng SUN. Genomic Characterization of a Recombinant Strain of PRRSV-2 between Lineages 1.8 and 1.5 [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(8): 3570-3578. |
| [10] | Huanqin ZHENG, Xiaomin JIANG, Hong YUE, Baoyan WANG, Yang LIU, Xingxiao ZHANG, Jianlong ZHANG, Hongwei ZHU. Isolation, Identification and Partial Biological Characteristics Analysis of Feline Herpesvirus-1 [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(7): 3040-3048. |
| [11] | Jitong LI, Tong ZHU, Junfeng LÜ, Yuehua GAO, Feng HU, Kexiang YU, Minxun SONG, Jianlin WANG, Yufeng LI. Isolation and Identification of Novel Picornavirus from Ducks and Whole Genome Sequence Analysis [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(7): 3075-3084. |
| [12] | Bohua LIU, Hanyu FU, Yuheng WANG, Suolangsizhu, Jiaqiang NIU, Yuhua BAO, Jiakui LI, Yefen XU. Isolation, Identification and Genome Analysis of Type B Pasteurella multocida Isolated from Yak in Tibetan Nakchu City [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(7): 3105-3118. |
| [13] | ZHENG Rui, LIU Zishi, ZHANG Kangyou, YAN Yong, WEI Ling, ZEREN Wengmu, DINGZE Demi, HUANG Jianjun, WANG Li, WEI Yong. Isolation, Identification and Biological Characterization of Colletotrichum jasminigenum in Stems of Peanuts [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(5): 2206-2213. |
| [14] | TIAN Rui, XU Sixiang, XIE Feng, LIU Guangjin, WANG Gang, LI Qingxia, DAI Lei, XIE Guoxin, ZHANG Qiongwen, LU Yajing, WANG Guangwen, WANG Jinxiu, ZHANG Wei. Bioinformatics Analysis of the Genome of Clostridium perfringens Isolated from Cattle [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(4): 1707-1715. |
| [15] | ZHOU Zhiyu, DU Jige, XIN Ruolan, ZHANG Jiawen, PAN Chenfan, YIN Chunsheng, CHEN Xiaoyun, ZHU Zhen. Isolation and Identification of Three Lumpy Skin Disease Viruses in China and Their GPCR Gene Analysis [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(12): 5620-5630. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||