





Acta Veterinaria et Zootechnica Sinica ›› 2025, Vol. 56 ›› Issue (9): 4241-4252.doi: 10.11843/j.issn.0366-6964.2025.09.010
• Review • Previous Articles Next Articles
LI Mengfan1( ), LI Qingyang1(
), LI Qingyang1( ), SONG Yanwen1(
), SONG Yanwen1( ), SONG Zhenhui1,2, ZHANG Xingcui1,*(
), SONG Zhenhui1,2, ZHANG Xingcui1,*( )
)
Received:2024-11-01
Online:2025-09-23
Published:2025-09-30
Contact:
ZHANG Xingcui
E-mail:1285530053@qq.com;1852112998@qq.com;962060187@qq.com;zhangxc923@163.com
CLC Number:
LI Mengfan, LI Qingyang, SONG Yanwen, SONG Zhenhui, ZHANG Xingcui. Structure and Function of Coronavirus S Proteins[J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(9): 4241-4252.
Table 1
Functional receptors and co-receptor factors of coronavirus"
| 属 Genera | 病毒 Viruses | 宿主 Hosts | 功能性受体 Functional receptors | 辅助性受体 Helper receptors | 参考文献 References |
| 甲型冠状病毒属 Alphacoronaviruses | HCoV-229E | Human | APN | [ | |
| HCoV-NL63 | Porcine | ACE2 | HS | [ | |
| TGEV | APN | SA | [ | ||
| PEDV | APN | SA | [ | ||
| SADS-CoV | Feline | HS、SA | [ | ||
| FECV | APN | SA | [ | ||
| FIPV Ⅰ | [ | ||||
| FIPV Ⅱ | Canine | APN | |||
| CCoV Ⅰ | [ | ||||
| CCoV Ⅱ | APN | ||||
| 乙型冠状病毒属 Betacoronavirus | SARS-CoV | Human | ACE2 | [ | |
| SARS-CoV2 | ACE2 | [ | |||
| MERS-CoV | DPP4 | SA | [ | ||
| HCoV-HKU1 | TMPRSS2 | SA | [ | ||
| HCoV-OC43 | SA | [ | |||
| BCoV | Bovine | APN | [ | ||
| MHV | Murine | CEACAM1 | [ | ||
| 丙型冠状病毒属 Gammacoronavirus | IBV | Avian | SA | [ | |
| 丁型冠状病毒属 Deltacoronavirus | PDCoV | Porcine | APN | [ |
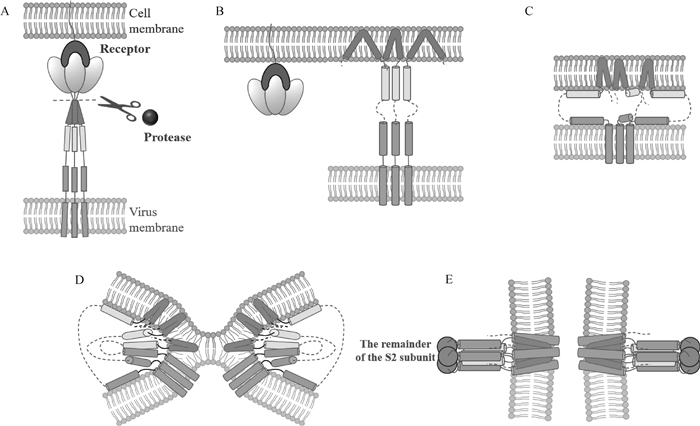
Fig. 4
The membrane fusion model of coronavirus S protein facilitates viral entry into cells[48](color images are provided in supplementary materials of OSID) After the S1 subunit binds to the receptor, the S protein is cleaved by protease (Fig. A), and the fusion peptide (FP) on the S2 subunit is exposed and penetrate into the host cell membrane (Fig. B). The heptad repeat 1 (HR1) and HR2 fold to form a six-helix bundle structure (Fig. C-E), aligning the FP with the single channel transmembrane anchor (TM), further promoting merging between the viral membrane and the cell membrane (Fig. D, E)"
| 1 |
VAKULENKO Y , DEVIATKIN A , DREXLER J F , et al. Modular evolution of coronavirus genomes[J]. Viruses, 2021, 13 (7): 1270.
doi: 10.3390/v13071270 |
| 2 |
HUANG C Y , DRACZKOWSKI P , WANG Y S , et al. In situ structure and dynamics of an alphacoronavirus spike protein by cryo-ET and cryo-EM[J]. Nat Commun, 2022, 13 (1): 4877.
doi: 10.1038/s41467-022-32588-3 |
| 3 |
SHANG J , ZHENG Y , YANG Y , et al. Cryo-EM structure of infectious bronchitis coronavirus spike protein reveals structural and functional evolution of coronavirus spike proteins[J]. PLoS Pathog, 2018, 14 (4): e1007009.
doi: 10.1371/journal.ppat.1007009 |
| 4 | LI Z , TOMLINSON A C , WONG A H , et al. The human coronavirus HCoV-229E S-protein structure and receptor binding[J]. Elife, 2019, 8, e1007009. |
| 5 |
WALLS A C , XIONG X , PARK Y J , et al. Unexpected receptor functional mimicry elucidates activation of coronavirus fusion[J]. Cell, 2019, 176 (5): 1026- 1039.e15.
doi: 10.1016/j.cell.2018.12.028 |
| 6 |
CHEN Y , ZHANG Z , LI J , et al. Porcine epidemic diarrhea virus S1 protein is the critical inducer of apoptosis[J]. Virol J, 2018, 15 (1): 170.
doi: 10.1186/s12985-018-1078-4 |
| 7 |
LI F . Receptor recognition mechanisms of coronaviruses: a decade of structural studies[J]. J Virol, 2015, 89 (4): 1954- 1964.
doi: 10.1128/JVI.02615-14 |
| 8 | WRAPP D , MCLELLAN J S . The 3.1-angstrom cryo-electron microscopy structure of the porcine epidemic diarrhea virus spike protein in the prefusion conformation[J]. J Virol, 2019, 93 (23): e00923- 19. |
| 9 |
YU J , QIAO S , GUO R , et al. Cryo-EM structures of HKU2 and SADS-CoV spike glycoproteins provide insights into coronavirus evolution[J]. Nat Commun, 2020, 11 (1): 3070.
doi: 10.1038/s41467-020-16876-4 |
| 10 | GUAN H , WANG Y , PER AČG ULIJA V , et al. Cryo-electron microscopy structure of the swine acute diarrhea syndrome coronavirus spike glycoprotein provides insights into evolution of unique coronavirus spike proteins[J]. J Virol, 2020, 94 (22): e01301- 20. |
| 11 |
WALLS A C , TORTORICI M A , BOSCH B J , et al. Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer[J]. Nature, 2016, 531 (7592): 114- 117.
doi: 10.1038/nature16988 |
| 12 |
WALLS A C , TORTORICI M A , FRENZ B , et al. Glycan shield and epitope masking of a coronavirus spike protein observed by cryo-electron microscopy[J]. Nat Struct Mol Biol, 2016, 23 (10): 899- 905.
doi: 10.1038/nsmb.3293 |
| 13 |
YANG T J , CHANG Y C , KO T P , et al. Cryo-EM analysis of a feline coronavirus spike protein reveals a unique structure and camouflaging glycans[J]. Proc Natl Acad Sci U S A, 2020, 117 (3): 1438- 1446.
doi: 10.1073/pnas.1908898117 |
| 14 |
ZHANG K , LI S , PINTILIE G , et al. A 3.4-Å cryo-electron microscopy structure of the human coronavirus spike trimer computationally derived from vitrified NL63 virus particles[J]. QRB Discov, 2020, 1, e11.
doi: 10.1017/qrd.2020.16 |
| 15 |
LI Y , WANG T , ZHANG J , et al. Exploring the regulatory function of the N-terminal domain of SARS-CoV-2 spike protein through molecular dynamics simulation[J]. Adv Theory Simul, 2021, 4 (10): 2100152.
doi: 10.1002/adts.202100152 |
| 16 |
SHANG J , WAN Y , LIU C , et al. Structure of mouse coronavirus spike protein complexed with receptor reveals mechanism for viral entry[J]. PLoS Pathog, 2020, 16 (3): e1008392.
doi: 10.1371/journal.ppat.1008392 |
| 17 |
LING A J W , CHANG L S , BABJI A S , et al. Review of sialic acid's biochemistry, sources, extraction and functions with special reference to edible bird's nest[J]. Food Chem, 2022, 367, 130755.
doi: 10.1016/j.foodchem.2021.130755 |
| 18 |
DIEP N V , NORIMINE J , SUEYOSHI M , et al. Novel porcine epidemic diarrhea virus (PEDV) variants with large deletions in the spike (S) gene coexist with PEDV strains possessing an intact S gene in domestic pigs in Japan: A new disease situation[J]. PLoS One, 2017, 12 (1): e0170126.
doi: 10.1371/journal.pone.0170126 |
| 19 |
PENG G , XU L , LIN Y L , et al. Crystal structure of bovine coronavirus spike protein lectin domain[J]. J Biol Chem, 2012, 287 (50): 41931- 41938.
doi: 10.1074/jbc.M112.418210 |
| 20 |
YOU R , LIU K , HUANG M , et al. Identification and comparison of the sialic acid-binding domain characteristics of avian coronavirus infectious bronchitis virus spike protein[J]. J Virol, 2023, 97 (5): e0048923.
doi: 10.1128/jvi.00489-23 |
| 21 |
QIAO M , LIN L , XIA K , et al. Recent advances in biotechnology for heparin and heparan sulfate analysis[J]. Talanta, 2020, 219, 121270.
doi: 10.1016/j.talanta.2020.121270 |
| 22 | NASKALSKA A , DABROWSKA A , SZCZEPANSKI A , et al. Membrane protein of human coronavirus NL63 is responsible for interaction with the adhesion receptor[J]. J Virol, 2019, 93 (19): e00355- 19. |
| 23 |
YANG Y L , WANG B , LI W , et al. Functional dissection of the spike glycoprotein S1 subunit and identification of cellular cofactors for regulation of swine acute diarrhea syndrome coronavirus entry[J]. J Virol, 2024, 98 (4): e0013924.
doi: 10.1128/jvi.00139-24 |
| 24 | DESMARETS L M B , THEUNS S , ROUKAERTS I D M , et al. Role of sialic acids in feline enteric coronavirus infections[J]. J Gen Virol, 2014, 95 (Pt 9): 1911- 1918. |
| 25 |
COOK S , CASTILLO D , WILLIAMS S , et al. Serotype Ⅰ and Ⅱ feline coronavirus replication and gene expression patterns of feline cells-building a better understanding of serotype I FIPV biology[J]. Viruses, 2022, 14 (7): 1356.
doi: 10.3390/v14071356 |
| 26 |
CERRACCHIO C , SERRA F , AMOROSO M G , et al. Canine coronavirus activates Aryl hydrocarbon receptor during in vitro infection[J]. Viruses, 2022, 14 (11): 2437.
doi: 10.3390/v14112437 |
| 27 | GUO H , HU B J , YANG X L , et al. Evolutionary arms race between virus and host drives genetic diversity in bat severe acute respiratory syndrome-related coronavirus spike genes[J]. J Virol, 2020, 94 (20): e00902- 20. |
| 28 |
LAN J , GE J , YU J , et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor[J]. Nature, 2020, 581 (7807): 215- 220.
doi: 10.1038/s41586-020-2180-5 |
| 29 |
ALAOFI A L . Exploring structural dynamics of the MERS-CoV receptor DPP4 and mutant DPP4 receptors[J]. J Biomol Struct Dyn, 2022, 40 (2): 752- 763.
doi: 10.1080/07391102.2020.1818626 |
| 30 |
SAUNDERS N , FERNANDEZ I , PLANCHAIS C , et al. TMPRSS2 is a functional receptor for human coronavirus HKU1[J]. Nature, 2023, 624 (7990): 207- 214.
doi: 10.1038/s41586-023-06761-7 |
| 31 |
TANG G , LIU Z , CHEN D . Human coronaviruses: Origin, host and receptor[J]. J Clin Virol, 2022, 155, 105246.
doi: 10.1016/j.jcv.2022.105246 |
| 32 |
JI W , PENG Q , FANG X , et al. Structures of a deltacoronavirus spike protein bound to porcine and human receptors[J]. Nat Commun, 2022, 13 (1): 1467.
doi: 10.1038/s41467-022-29062-5 |
| 33 |
SHI J , SHI Y , XIU R , et al. Identification of a novel neutralizing epitope on the N-terminal domain of the human coronavirus 229E spike protein[J]. J Virol, 2022, 96 (4): e0195521.
doi: 10.1128/jvi.01955-21 |
| 34 | SHANG J , ZHENG Y , YANG Y , et al. Cryo-electron microscopy structure of porcine deltacoronavirus spike protein in the prefusion state[J]. J Virol, 2018, 92 (4): e01556- 17. |
| 35 |
PAN Y , TIAN X , QIN P , et al. Discovery of a novel swine enteric alphacoronavirus (SeACoV) in southern China[J]. Vet Microbiol, 2017, 211, 15- 21.
doi: 10.1016/j.vetmic.2017.09.020 |
| 36 |
LI F . Structure, function, and evolution of coronavirus spike proteins[J]. Annu Rev Virol, 2016, 3 (1): 237- 261.
doi: 10.1146/annurev-virology-110615-042301 |
| 37 |
LI C , SU M , YIN B , et al. Integrin αvβ3 enhances replication of porcine epidemic diarrhea virus on Vero E6 and porcine intestinal epithelial cells[J]. Vet Microbiol, 2019, 237, 108400.
doi: 10.1016/j.vetmic.2019.108400 |
| 38 |
WARDEH M , BAYLIS M , BLAGROVE M S C . Predicting mammalian hosts in which novel coronaviruses can be generated[J]. Nat Commun, 2021, 12 (1): 780.
doi: 10.1038/s41467-021-21034-5 |
| 39 |
WANG S , XU C , SHI J , et al. Regulatory effect and mechanism of APN gene on porcine epidemic diarrhea virus resistance[J]. Gene, 2021, 775, 145448.
doi: 10.1016/j.gene.2021.145448 |
| 40 |
PIZZANELLI S , FORTE C , PINZINO C , et al. Copper(Ⅱ) complexes with peptides based on the second cell binding site of fibronectin: metal coordination and ligand exchange kinetics[J]. Phys Chem Chem Phys, 2016, 18 (5): 3982- 3994.
doi: 10.1039/C5CP05798A |
| 41 |
CASTILLO G , MORA-DíAZ J C , NELLI R K , et al. Human air-liquid-interface organotypic airway cultures express significantly more ACE2 receptor protein and are more susceptible to HCoV-NL63 infection than monolayer cultures of primary respiratory epithelial cells[J]. Microbiol Spectr, 2022, 10 (4): e0163922.
doi: 10.1128/spectrum.01639-22 |
| 42 | PECK K M , SCOBEY T , SWANSTROM J , et al. Permissivity of dipeptidyl peptidase 4 orthologs to Middle East respiratory syndrome coronavirus is governed by glycosylation and other complex determinants[J]. J Virol, 2017, 91 (19): e00534- 17. |
| 43 | PALLESEN J , WANG N , CORBETT K S , et al. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen[J]. Proc Natl Acad Sci U S A, 2017, 114 (35): E7348- E7357. |
| 44 |
KIRCHDOERFER R N , WANG N , PALLESEN J , et al. Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis[J]. Sci Rep, 2018, 8 (1): 15701.
doi: 10.1038/s41598-018-34171-7 |
| 45 |
GUI M , SONG W , ZHOU H , et al. Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding[J]. Cell Res, 2017, 27 (1): 119- 129.
doi: 10.1038/cr.2016.152 |
| 46 |
WALLS A C , TORTORICI M A , SNIJDER J , et al. Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion[J]. Proc Natl Acad Sci U S A, 2017, 114 (42): 11157- 11162.
doi: 10.1073/pnas.1708727114 |
| 47 |
SONG X , SHI Y , DING W , et al. Cryo-EM analysis of the HCoV-229E spike glycoprotein reveals dynamic prefusion conformational changes[J]. Nat Commun, 2021, 12 (1): 141.
doi: 10.1038/s41467-020-20401-y |
| 48 |
ALIPER E T , EFREMOV R G . Inconspicuous yet indispensable: The coronavirus spike transmembrane domain[J]. Int J Mol Sci, 2023, 24 (22): 16421.
doi: 10.3390/ijms242216421 |
| 49 |
KIELIAN M . Mechanisms of virus membrane fusion proteins[J]. Annu Rev Virol, 2014, 1 (1): 171- 189.
doi: 10.1146/annurev-virology-031413-085521 |
| 50 |
SHI W , CAI Y , ZHU H , et al. Cryo-EM structure of SARS-CoV-2 postfusion spike in membrane[J]. Nature, 2023, 619 (7969): 403- 409.
doi: 10.1038/s41586-023-06273-4 |
| 51 |
BASSO L G M , ZERAIK A E , FELIZATTI A P , et al. Membranotropic and biological activities of the membrane fusion peptides from SARS-CoV spike glycoprotein: The importance of the complete internal fusion peptide domain[J]. Biochim Biophys Acta Biomembr, 2021, 1863 (11): 183697.
doi: 10.1016/j.bbamem.2021.183697 |
| 52 |
WALLS A C , PARK Y J , TORTORICI M A , et al. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein[J]. Cell, 2020, 181 (2): 281- 92.e6.
doi: 10.1016/j.cell.2020.02.058 |
| 53 |
MAEDA D , TIAN D , YU H , et al. Killed whole-genome reduced-bacteria surface-expressed coronavirus fusion peptide vaccines protect against disease in a porcine model[J]. Proc Natl Acad Sci U S A, 2021, 118 (18): e2025622118.
doi: 10.1073/pnas.2025622118 |
| 54 |
DACON C , TUCKER C , PENG L , et al. Broadly neutralizing antibodies target the coronavirus fusion peptide[J]. Science, 2022, 377 (6607): 728- 735.
doi: 10.1126/science.abq3773 |
| 55 |
LOW J S , JERAK J , TORTORICI M A , et al. ACE2-binding exposes the SARS-CoV-2 fusion peptide to broadly neutralizing coronavirus antibodies[J]. Science, 2022, 377 (6607): 735- 742.
doi: 10.1126/science.abq2679 |
| 56 |
SUN X , YI C , ZHU Y , et al. Neutralization mechanism of a human antibody with pan-coronavirus reactivity including SARS-CoV-2[J]. Nat Microbiol, 2022, 7 (7): 1063- 1074.
doi: 10.1038/s41564-022-01155-3 |
| 57 |
MILLET J K , WHITTAKER G R . Host cell proteases: Critical determinants of coronavirus tropism and pathogenesis[J]. Virus Res, 2015, 202, 120- 34.
doi: 10.1016/j.virusres.2014.11.021 |
| 58 | YAN L , MENG B , XIANG J , et al. Crystal structure of the post-fusion core of the human coronavirus 229E spike protein at 1.86 Å resolution[J]. Acta Crystallogr D Struct Biol, 2018, 74 (Pt 9): 841- 851. |
| 59 |
WANG F , YANG G , YAN L . Crystal Structures of Fusion Cores from CCoV-HuPn-2018 and SADS-CoV[J]. Viruses, 2024, 16 (2): 272.
doi: 10.3390/v16020272 |
| 60 |
WESTERFIELD J M , BARRERA F N . Membrane receptor activation mechanisms and transmembrane peptide tools to elucidate them[J]. J Biol Chem, 2020, 295 (7): 1792- 1814.
doi: 10.1074/jbc.REV119.009457 |
| 61 |
KUMAR P , BHARDWAJ T , GARG N , et al. Microsecond simulations and CD spectroscopy reveals the intrinsically disordered nature of SARS-CoV-2 spike-C-terminal cytoplasmic tail (residues 1242-1273) in isolation[J]. Virology, 2022, 566, 42- 55.
doi: 10.1016/j.virol.2021.11.005 |
| 62 |
SADASIVAN J , SINGH M , SARMA J D . Cytoplasmic tail of coronavirus spike protein has intracellular targeting signals[J]. J Biosci, 2017, 42 (2): 231- 244.
doi: 10.1007/s12038-017-9676-7 |
| 63 |
WU Z , ZHANG Z , WANG X , et al. Palmitoylation of SARS-CoV-2 S protein is essential for viral infectivity[J]. Signal Transduct Target Ther, 2021, 6 (1): 231.
doi: 10.1038/s41392-021-00651-y |
| 64 |
LUO Y , TAN C W , XIE S Z , et al. Identification of ZDHHC17 as a potential drug target for swine acute diarrhea syndrome coronavirus infection[J]. mBio, 2021, 12 (5): e0234221.
doi: 10.1128/mBio.02342-21 |
| 65 |
GELHAUS S , THAA B , ESCHKE K , et al. Palmitoylation of the Alphacoronavirus TGEV spike protein S is essential for incorporation into virus-like particles but dispensable for S-M interaction[J]. Virology, 2014, 464-465, 397- 405.
doi: 10.1016/j.virol.2014.07.035 |
| 66 |
KUMAR P , BHARDWAJ A , MUKHERJEE B , et al. Coronaviruses spike glycoprotein endodomains: The sequence and structure-based comprehensive study[J]. Protein Sci, 2023, 32 (11): e4804.
doi: 10.1002/pro.4804 |
| 67 |
CHENG Y R , LI X , ZHAO X , et al. Cell entry of animal coronaviruses[J]. Viruses, 2021, 13 (10): 1977.
doi: 10.3390/v13101977 |
| 68 |
JOHNSON B A , XIE X , BAILEY A L , et al. Loss of furin cleavage site attenuates SARS-CoV-2 pathogenesis[J]. Nature, 2021, 591 (7849): 293- 299.
doi: 10.1038/s41586-021-03237-4 |
| 69 |
WANG Z , ZHONG K , WANG G , et al. Loss of furin site enhances SARS-CoV-2 spike protein pseudovirus infection[J]. Gene, 2023, 856, 147144.
doi: 10.1016/j.gene.2022.147144 |
| 70 | ZEHR J D , KOSAKOVSKY POND S L , MILLET J K , et al. Natural selection differences detected in key protein domains between non-pathogenic and pathogenic feline coronavirus phenotypes[J]. Virus Evol, 2023, 9 (1): 523607. |
| 71 |
KIM J , YOON J , PARK J E . Furin cleavage is required for swine acute diarrhea syndrome coronavirus spike protein-mediated cell-cell fusion[J]. Emerg Microbes Infect, 2022, 11 (1): 2176- 2183.
doi: 10.1080/22221751.2022.2114850 |
| 72 |
BONNIN A , DANNEELS A , DUBUISSON J , et al. HCoV-229E spike protein fusion activation by trypsin-like serine proteases is mediated by proteolytic processing in the S2' region[J]. J Gen Virol, 2018, 99 (7): 908- 912.
doi: 10.1099/jgv.0.001074 |
| 73 |
LIN F , ZHANG H , LI L , et al. PEDV: Insights and advances into types, function, structure, and receptor recognition[J]. Viruses, 2022, 14 (8): 1744.
doi: 10.3390/v14081744 |
| 74 |
WICHT O , LI W , WILLEMS L , et al. Proteolytic activation of the porcine epidemic diarrhea coronavirus spike fusion protein by trypsin in cell culture[J]. J Virol, 2014, 88 (14): 7952- 7961.
doi: 10.1128/JVI.00297-14 |
| 75 |
SUN M , MA J , YU Z , et al. Identification of two mutation sites in spike and envelope proteins mediating optimal cellular infection of porcine epidemic diarrhea virus from different pathways[J]. Vet Res, 2017, 48 (1): 44.
doi: 10.1186/s13567-017-0449-y |
| [1] | CHANG Shuo, SUN Xiuzhu, REN Zhanjun, WANG Shuhui. Research Progress in Rabbit Genomics [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(8): 3578-3590. |
| [2] | LIU Sha, YANG Caichun, ZHANG Xiaoyu, CHEN Qiong, LIU Xiong, CHEN Hongbo, ZHOU Huanhuan, SHI Liangyu. Population Genetic Structure and Genome-wide Runs of Homozygosity Analysis in Meihuaxing Pigs Based on 80K SNP Chip [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(8): 3749-3760. |
| [3] | REN Qianzi, ZHANG Baizhong, WANG Zhenqing, WANG Xianglin, GONG Ying, HU Renke, PU Yabin, SU Peng, LI Yefang, MA Yuehui, LI Haobang, JIANG Lin. Genetic Evolutionary Analysis of Wuxue Goat Based on Whole Genome Resequencing [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(8): 3787-3801. |
| [4] | LI Qian, GAO Huan, FU Shuang, SUO Zhuo, DAI Yue, CHEN Chen, LI Rongtian, LENG Jing. Anaerobic Fungi of Digestive Tract and Their Interactions with Other Microorganisms [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(7): 3096-3106. |
| [5] | XIANG Lingxian, JI Qianyu, SHAN Xinxin, LI Lin. Advances in the Study of Drug Resistance and Pathogenicity of Bacterial Two-component Systems [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(7): 3116-3128. |
| [6] | ZHANG Jialiang, HUANG Chang, YANG Yonglin, YANG Hua, BAI Wenlin, MA Yuehui, ZHAO Qianjun. Genetic Structure and Wool Trait Selection Signatures Analysis of Chinese Sheep Populations Based on 50K Liquid SNP Chip [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(7): 3164-3176. |
| [7] | DONG Jiaojiao, DING Hong, ZHANG Yinliang, ZHANG Ran, LIU Huage, ZANG Sumin, ZHANG Zhenhong, ZHOU Rongyan, LI Lanhui. Differences and Functional Analysis of Cecal Flora in Taihang Chickens Infected with Salmonella Pullorum [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(6): 2741-2751. |
| [8] | WANG Qinqian, GAO Zhendong, LU Ying, MA Ruoshan, DENG Weidong, HE Xiaoming. Research Progress of Whole Genome Resequencing in Chinese Indigenous Cattle [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(5): 2026-2037. |
| [9] | QIN Xiaoxia, GAN Haiqing, SHE Gaojin, LIU Yong, HUANG Xingguo, CHEN Lirong, YANG Lingyuan. Research Progess in Active Components, Biological Functions of Camellia Seed Meal and Its Applications in Livestock and Poultry Production [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(5): 2070-2081. |
| [10] | CHEN Ting, CUI Yadong, LAN Wei, KONG Xiangfeng. Function of Glucosamine and Its Application in Animal Production [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(4): 1518-1526. |
| [11] | ZHU Yun, WANG Yuming, SUN Xiaoxiao, CHEN Hui, ZHAO Feng, XIE Jingjing, CHEN Yifan, SA Renna. Effect of the Addition of Corn Gluten Meal to Low-protein Diversified Diet on Growth Performance and Digestive Characteristics of White-feathered Broilers [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(4): 1802-1812. |
| [12] | LI Xiaotong, WANG Pengyu, FANG Yingyan, YU Hongxi, ZHANG Yi, WANG Yachun, ZHANG Yuanpei, LI Yanqin, JIANG Li. Mining and Functional Verification of Gene Polymorphisms Loci Related to Bull Sperm Freezability [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(4): 1981-1988. |
| [13] | ZHANG Yanmin, LIU Shuai, TENG Zhanwei, XIE Hongbing, XIA Xiaojing, HE Yonghui, CHANG Meinan. Research Progress on the Mechanism of Functional Oligosaccharides Alleviating Calf Diarrhea [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(3): 979-994. |
| [14] | WANG Haoyu, MA Keyan, LI Taotao, LI Dengpan, ZHAO Qing, MA Youji. Population Genetic Diversity and Population Structure Analysis of Small-boned Goat Based on Specific-Locus Amplified Fragment Sequencing [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(3): 1170-1179. |
| [15] | HU Xin, YOU Wei, JIANG Fugui, CHENG Haijian, SUN Zhigang, SONG Enliang. Analysis of Genetic Diversity and Population Structure of Simmental Cattle Based on Whole Genome Resequencing [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(3): 1189-1202. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||