





Acta Veterinaria et Zootechnica Sinica ›› 2025, Vol. 56 ›› Issue (10): 4947-4962.doi: 10.11843/j.issn.0366-6964.2025.10.016
• Animal Genetics and Breeding • Previous Articles Next Articles
FENG Da( ), WEI Chen*(
), WEI Chen*( ), HU Siyi, DU Chunmei, MA Jian, WU Jiang, ZHOU Guangxian, GAN Shangquan*(
), HU Siyi, DU Chunmei, MA Jian, WU Jiang, ZHOU Guangxian, GAN Shangquan*( )
)
Received:2025-03-13
Online:2025-10-23
Published:2025-11-01
Contact:
WEI Chen, GAN Shangquan
E-mail:fda_1020@163.com;weichenwjf@126.com;shangquangan@gdou.edu.cn
CLC Number:
FENG Da, WEI Chen, HU Siyi, DU Chunmei, MA Jian, WU Jiang, ZHOU Guangxian, GAN Shangquan. Genetic Diversity and Population Structure Analysis of Leizhou Goat Based on Whole Genome Resequencing Analysis[J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(10): 4947-4962.

Fig. 1
The distribution of SNPs in Leizhou goat chromosomes and their genomic regions annotation A. The distribution of SNPs on 29 chromosomes, with the y-axis representing 1-29 chromosomes and the x-axis representing every 1 Mb window on the chromosome. Different colors indicate the number of SNPs contained in each 1 Mb window. B. Annotation status of SNPs in genomic regions"
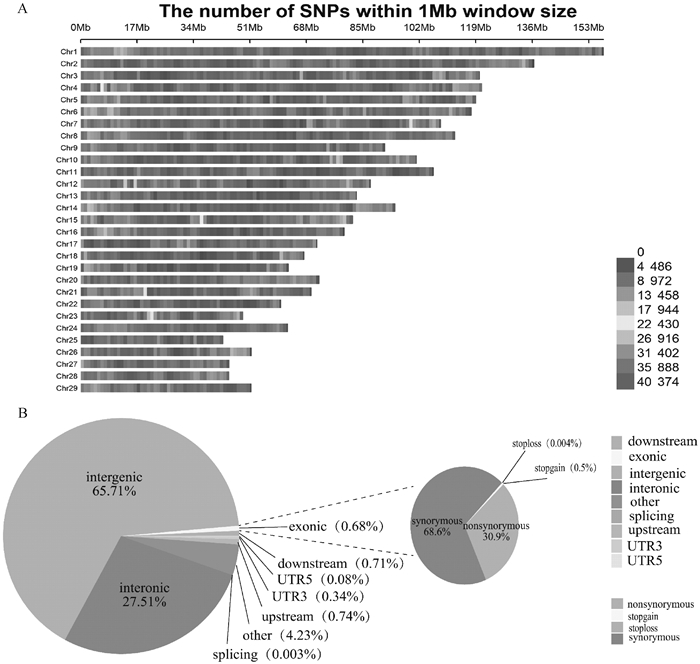
Table 1
Genetic diversity indexes of 6 goat populations"
| 群体 Population | 最小等位基因频率 MAF | 观测杂合度 HO | 期望杂合度 HE | 有效群体含量 Ne | 多态信息含量 PIC | 核苷酸多态性 Pi | 近交系数 FIS |
| 雷州山羊 LZ | 0.211 | 0.298 | 0.295 | 65.488 | 0.808 | 1.798×10-3 | 0.086 |
| 云上黑山羊 YS | 0.207 | 0.305 | 0.290 | 21.312 | 0.938 | 2.179×10-3 | 0.053 |
| 金堂黑山羊 JT | 0.198 | 0.280 | 0.281 | 23.478 | 0.845 | 1.730×10-3 | 0.092 |
| 济宁青山羊 JN | 0.188 | 0.276 | 0.269 | 24.300 | 0.947 | 2.354×10-3 | 0.299 |
| 西藏山羊 TB | 0.188 | 0.250 | 0.270 | 25.879 | 0.947 | 2.080×10-3 | 0.083 |
| 隆林山羊 LL | 0.227 | 0.313 | 0.315 | 16.124 | 0.750 | 2.043×10-3 | 0.075 |

Fig. 2
Linkage disequilibrium(LD)decay estimated from the 6 goat populations The horizontal coordinate represents the physical distance (kb), and the vertical coordinate represents the LD coefficient (r2). LZ. Leizhou goat; LL. Longlin goat; TB. Tibetan goat; JN. Jining grey goat; JT. Jintang black goat; YS. Yunshang black goat, the same as below"
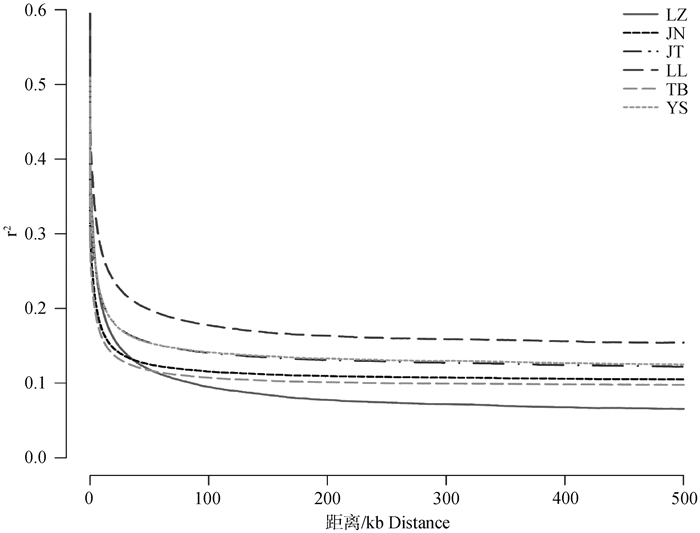

Fig. 3
ROH analysis in 6 goat populations A. The distribution of ROH number and length for each individual, with each point representing an individual and the color of the point indicating the population to which the individual belongs. The horizontal axis corresponds to the ROH number for each individual, and the vertical axis corresponds to the ROH length (Mb) for each individual. B. The violin plot of FROH distribution, with the horizontal axis representing each population and the vertical axis representing the FROH coefficient. The width of the violin represents the distribution of individuals, and the box part of the internal boxplot represents the quartile range, with the horizontal line inside the box representing the median"
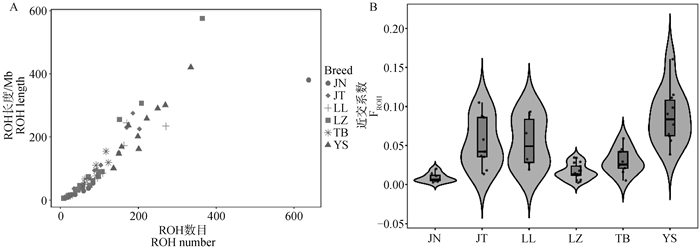

Fig. 4
The visualization results of G matrix analysis for 6 goat populations The horizontal and vertical coordinates are goat individuals, each small square represents the genetic relationship between two individuals, the closer the color is to red, the closer the genetic relationship between two individuals, and vice versa"
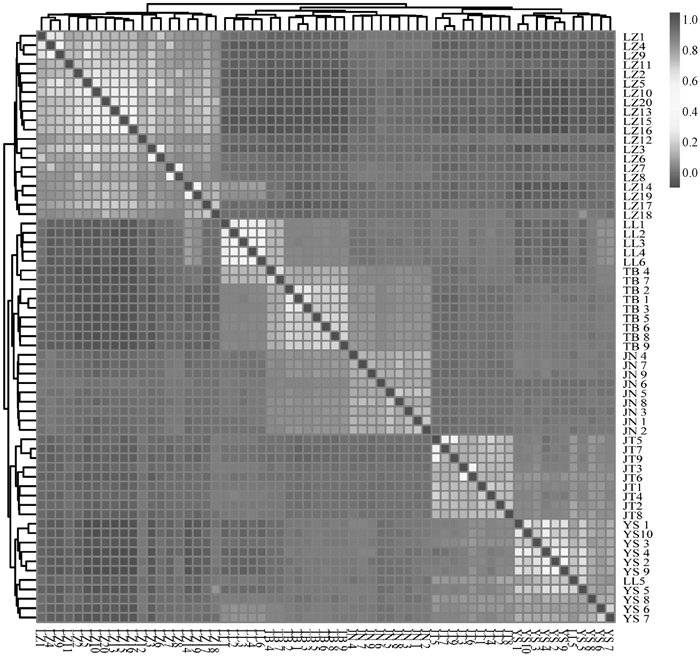

Fig. 5
The results of IBS genetic distance matrix for 6 goat populations The horizontal and vertical coordinates are goat individuals, each small square represents the genetic distance value between two individuals, the closer the color is to red, the greater the genetic distance between two individuals, and vice versa"
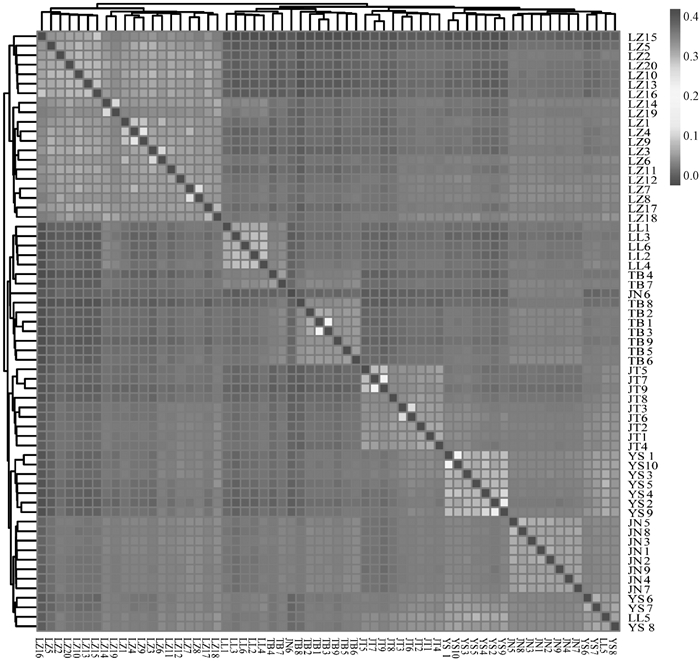
Fig. 7
FST analysis of goat populations A. FST density plot, where the x-axis represents the FST index, and different colors indicate comparisons between different populations. B. FST heatmap, where color intensity indicates the magnitude of the FST index, and each square represents the average FST value between the corresponding population on the x-axis and the population on the y-axis. C. Manhattan plot of FST analysis between the LZ and TB populations. D. Manhattan plot of FST analysis between the LZ and YS populations"
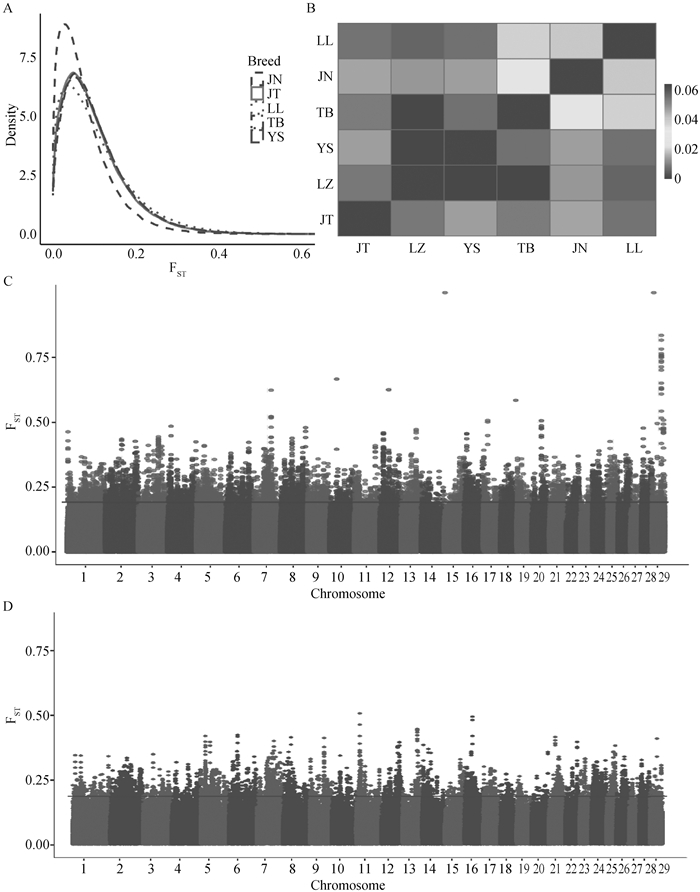
Table 2
GO functional enrichment analysis of LZ and TB populations"
| GO条目 GO term | P值 Pvalue | 基因 Gene |
| olfactory receptor activity | 1.9×10-4 | LOC102185414, LOC102183215, LOC102183415, LOC102182462, LOC102184860, LOC102183344等 |
| G protein-coupled receptor activity | 2.8×10-4 | ADGRL3, LOC102185414, LOC102183215, LOC102183415, LOC102182462, LOC102184860等 |
| G protein-coupled receptor signaling pathway | 7.1×10-4 | CXCR1, LOC102185414, LOC102183215, LOC102183415, LOC102182462, LOC102184860等 |
| plasma membrane | 0.001 1 | ADGRL3, SHC2, GRIA4, CRIPTO, NPC1, DNAJC5等 |
| odorant binding | 0.002 0 | LOC102185414, LOC102183215, LOC102175834, LOC102169980, LOC102182462, LOC102184860 |
| sensory perception of smell | 0.002 2 | LOC102185414, LOC102183215, LOC102175834, LOC102169980, LOC102182462, LOC102184860 |
| lysozyme activity | 0.003 9 | LOC102185900, LOC102172331, LOC108633237 |
| defense response to Gram-negative bacterium | 0.004 3 | LOC102171106, LOC102185900, LOC102172331, LOC108633237 |
| killing of cells of another organism | 0.009 9 | LOC102185900, LOC102172331, LOC108633237 |

Fig. 8
KEGG pathways analysis map of LZ, TB and YS populations A. KEGG pathway diagram of significantly enriched candidate genes identified by FST analysis between LZ and TB populations. B. KEGG pathway diagram of significantly enriched candidate genes identified by FST analysis between LZ and YS populations. The y-axis represents KEGG enrichment pathway names, the x-axis represents the enrichment score. The size of the circles corresponds to the number of enriched genes; The color gradient of the circles indicates the significance level"
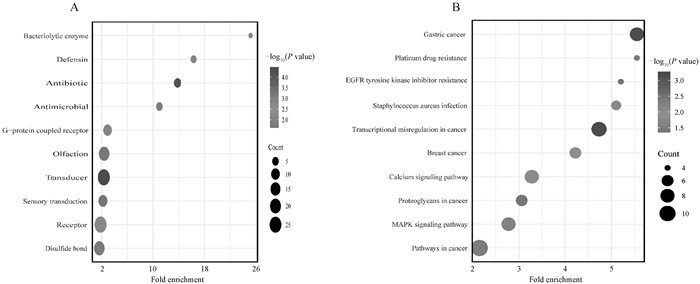
Table 3
GO functional enrichment analysis of LZ and YS populations"
| GO条目 GO term | P值 P value | 基因 Gene |
| G protein-coupled receptor signaling pathway | 1.1×10-6 | LOC102187858, LOC102182402, LOC102183415, LOC108634193, LOC102186762, LOC102187045等 |
| olfactory receptor activity | 1.9×10-6 | LOC102187858, LOC102182402, LOC102183415, LOC108634193, LOC102186762, LOC102187045等 |
| membrane | 1.2×10-5 | CEBPZOS, UPK3BL2, NUDT4, SEC11C, TMEM254, BST2, NAT8, DNAJC5, OSBPL1A等 |
| G protein-coupled receptor activity | 1.5×10-5 | LOC102187858, LOC102182402, LOC102183415, LOC108634193, LOC102186762, LOC102187045等 |
| structural constituent of eye lens | 0.005 9 | CRYGC, LOC102191000, LOC102188265 |
| lens development in camera-type eye | 0.011 | CRYGC, LOC102191000, LOC102188265 |
| cyanamide hydratase activity | 0.019 | LOC106502860, LOC108637506 |
| plasma membrane | 0.028 | HTR1F, SHC2, PIEZO2, GRIA4, SYTL2, DNAJC5, OSBPL1A |
| Glycerophosphodiester phosphodiesterase activity | 0.031 | LOC102183767, LOC102183296 |
| lipid transport | 0.047 | OSBPL1A, LOC102172041, LOC102172606 |
| 1 | 国家畜禽遗传资源委员会.中国畜禽遗传资源志(羊志)[M].北京:中国农业出版社,2011. |
| China National Commission of Animal Genetic Resources.Animal genetic resources in China(Sheep and Goats)[M].Beijing:China Agriculture Press,2011. | |
| 2 |
KICHAMUN,WANJALAG,CZISZTERL T,et al.Assessing the population structure and genetic variability of kenyan native goats under extensive production system[J].Sci Rep,2024,14(1):16342.
doi: 10.1038/s41598-024-67374-2 |
| 3 |
MUKHINAV,SVISHCHEVAG,VORONKOVAV,et al.Genetic diversity, population structure and phylogeny of indigenous goats of mongolia revealed by SNP genotyping[J].Animals,2022,12(3):221.
doi: 10.3390/ani12030221 |
| 4 |
VISSERC,LASHMARS F,VAN MARLE-KOSTERE,et al.Genetic diversity and population structure in south african, french and argentinian angora goats from genome-wide SNP data[J].PLoS ONE,2016,11(5):e0154353.
doi: 10.1371/journal.pone.0154353 |
| 5 |
VAN DIJKE L,AUGERH,JASZCZYSZYNY,et al.Ten years of next-generation sequencing technology[J].Trends Genet,2014,30(9):418-426.
doi: 10.1016/j.tig.2014.07.001 |
| 6 | GOODWINS,MCPHERSONJ D,MCCOMBIEW R.Coming of age: Ten years of next-generation sequencing technologies[J].Nat Rev Genet,2016,17(6):333-351. |
| 7 | SHASTRYB S.SNPs: Impact on Gene Function and Phenotype[J].Methods Mol Biol,2009,578,3-22. |
| 8 |
ZHUZ,ZHANGL,XINQ,et al.Whole-genome sequencing revealed the population structure of fujian chicken breeds[J].Czech J Anim Sci,2024,69(8):323-330.
doi: 10.17221/91/2023-CJAS |
| 9 |
TONGX,HOUL,HEW,et al.Whole genome sequence analysis reveals genetic structure and X-chromosome haplotype structure in indigenous Chinese pigs[J].Sci Rep,2020,10(1):9433.
doi: 10.1038/s41598-020-66061-2 |
| 10 |
ZHANGY,WEIZ,ZHANGM,et al.Population structure and selection signal analysis of nanyang cattle based on whole-genome sequencing data[J].Genes,2024,15(3):351.
doi: 10.3390/genes15030351 |
| 11 |
SHIH,LIT,SUM,et al.Whole genome sequencing revealed genetic diversity, population structure, and selective signature of panou tibetan sheep[J].BMC Genomics,2023,24(1):50.
doi: 10.1186/s12864-023-09146-2 |
| 12 |
CHANGL,ZHENGY,LIS,et al.Identification of genomic characteristics and selective signals in guizhou black goat[J].BMC Genomics,2024,25(1):164.
doi: 10.1186/s12864-023-09954-6 |
| 13 |
CHENQ,HUANGY,WANGZ,et al.Whole-genome resequencing reveals diversity and selective signals in Longlin goat[J].Gene,2021,771,145371.
doi: 10.1016/j.gene.2020.145371 |
| 14 |
EG X,ZHAOY J,CHENL P,et al.Genetic diversity of the Chinese goat in the littoral zone of the yangtze river as assessed by microsatellite and mtDNA[J].Ecol Evol,2018,8(10):5111-5123.
doi: 10.1002/ece3.4100 |
| 15 | 郭家中,张一菲,武国,等.利用全基因组重测序分析雅安奶山羊遗传多样性和遗传结构[J].四川农业大学学报,2025,43(2):444-451. |
| GUOJ Z,ZHANGY F,WUG,et al.Analysis on genetic diversity and genetic structure of Yaan dairy goats based on whole-genome resequencing[J].Journal of Sichuan Agricultural University,2025,43(2):444-451. | |
| 16 |
MAK,LID,QIX,et al.Population structure, runs of homozygosity analysis and construction of single nucleotide polymorphism fingerprinting database of Longnan goat population[J].Food Energy Secur,2024,13(1):e517.
doi: 10.1002/fes3.517 |
| 17 |
SHERIFFO,AHBARAA M,HAILEA,et al.Whole-genome resequencing reveals genomic variation and dynamics in Ethiopian indigenous goats[J].Front Genet,2024,15,1353026.
doi: 10.3389/fgene.2024.1353026 |
| 18 |
CHENS,ZHOUY,CHENY,et al.fastp: An ultra-fast all-in-one FASTQ preprocessor[J].Bioinformatics,2018,34(17):i884-i890.
doi: 10.1093/bioinformatics/bty560 |
| 19 |
LIH,DURBINR.Fast and accurate short read alignment with burrows-wheeler transform[J].Bioinformatics,2009,25(14):1754-1760.
doi: 10.1093/bioinformatics/btp324 |
| 20 |
BICKHARTD M,ROSENB D,KORENS,et al.Single-molecule sequencing and chromatin conformation capture enable de novo reference assembly of the domestic goat genome[J].Nat Genet,2017,49(4):643-650.
doi: 10.1038/ng.3802 |
| 21 |
LIH,HANDSAKERB,WYSOKERA,et al.The sequence alignment/map format and SAMtools[J].Bioinformatics,2009,25(16):2078-2079.
doi: 10.1093/bioinformatics/btp352 |
| 22 |
MCKENNAA,HANNAM,BANKSE,et al.The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data[J].Genome Res,2010,20(9):1297-1303.
doi: 10.1101/gr.107524.110 |
| 23 |
PEDERSENB S,QUINLANA R.Mosdepth: Quick coverage calculation for genomes and exomes[J].Bioinformatics,2018,34(5):867-868.
doi: 10.1093/bioinformatics/btx699 |
| 24 |
YANGH,WANGK.Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR[J].Nat Protoc,2015,10(10):1556-1566.
doi: 10.1038/nprot.2015.105 |
| 25 |
PURCELLS,NEALEB,TODD-BROWNK,et al.PLINK: A tool set for whole-genome association and population-based linkage analyses[J].Am J Hum Genet,2007,81(3):559-575.
doi: 10.1086/519795 |
| 26 |
DANECEKP,AUTONA,ABECASISG,et al.The variant call format and VCFtools[J].Bioinformatics,2011,27(15):2156-2158.
doi: 10.1093/bioinformatics/btr330 |
| 27 |
ZHANGC,DONGS S,XUJ Y,et al.PopLDdecay: A fast and effective tool for linkage disequilibrium decay analysis based on variant call format files[J].Bioinformatics,2019,35(10):1786-1788.
doi: 10.1093/bioinformatics/bty875 |
| 28 |
YANGJ,LEES H,GODDARDM E,et al.GCTA: A tool for genome-wide complex trait analysis[J].Am J Hum Genet,2011,88(1):76-82.
doi: 10.1016/j.ajhg.2010.11.011 |
| 29 |
YANGJ,BENYAMINB,MCEVOYB P,et al.Common SNPs explain a large proportion of the heritability for human height[J].Nat Genet,2010,42(7):565-569.
doi: 10.1038/ng.608 |
| 30 | VILLANUEVAR A M,CHENZ J.ggplot2: Elegant graphics for data analysis (2nd ed.)[J].Meas-Interdiscip Res,2019,17(3):160-167. |
| 31 |
LEET H,GUOH,WANGX,et al.SNPhylo: a pipeline to construct a phylogenetic tree from huge SNP data[J].BMC Genomics,2014,15(1):162.
doi: 10.1186/1471-2164-15-162 |
| 32 |
ALEXANDERD H,NOVEMBREJ,LANGEK.Fast model-based estimation of ancestry in unrelated individuals[J].Genome Res,2009,19(9):1655-1664.
doi: 10.1101/gr.094052.109 |
| 33 |
QUINLANA R,HALLI M.BEDTools: A flexible suite of utilities for comparing genomic features[J].Bioinformatics,2010,26(6):841-842.
doi: 10.1093/bioinformatics/btq033 |
| 34 |
SHERMANB T,HAOM,QIUJ,et al.DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update)[J].Nucleic Acids Res,2022,50(W1):W216-W221.
doi: 10.1093/nar/gkac194 |
| 35 |
COLLIL,MILANESIM,TALENTIA,et al.Genome-wide SNP profiling of worldwide goat populations reveals strong partitioning of diversity and highlights post-domestication migration routes[J].Genet Sel Evol,2018,50(1):58.
doi: 10.1186/s12711-018-0422-x |
| 36 |
SUNX,GUOJ,LIL,et al.Genetic diversity and selection signatures in jianchang black goats revealed by whole-genome sequencing data[J].Animals,2022,12(18):2365.
doi: 10.3390/ani12182365 |
| 37 |
ZHANGT,WANGZ,LIY,et al.Genetic diversity and population structure in five inner mongolia cashmere goat populations using whole-genome genotyping[J].Anim Biosci,2024,37(7):1168-1176.
doi: 10.5713/ab.23.0424 |
| 38 |
GEBRESELASEH B,NIGUSSIEH,WANGC,et al.Genetic diversity, population structure and selection signature in begait goats revealed by whole-genome sequencing[J].Animals,2024,14(2):307.
doi: 10.3390/ani14020307 |
| 39 |
WEIC,LUJ,XUL,et al.Genetic structure of Chinese indigenous goats and the special geographical structure in the southwest China as a geographic barrier driving the fragmentation of a large population[J].PLoS ONE,2014,9(4):e94435.
doi: 10.1371/journal.pone.0094435 |
| 40 |
CAIY,FUW,CAID,et al.Ancient genomes reveal the evolutionary history and origin of cashmere-producing goats in China[J].Mol Biol Evol,2020,37(7):2099-2109.
doi: 10.1093/molbev/msaa103 |
| 41 |
CEBALLOSF C,JOSHIP K,CLARKD W,et al.Runs of homozygosity: Windows into population history and trait architecture[J].Nat Rev Genet,2018,19(4):220-234.
doi: 10.1038/nrg.2017.109 |
| 42 |
KELLERM C,VISSCHERP M,GODDARDM E.Quantification of inbreeding due to distant ancestors and its detection using dense single nucleotide polymorphism data[J].Genetics,2011,189(1):237-249.
doi: 10.1534/genetics.111.130922 |
| 43 |
HUANGC,ZHAOQ,CHENQ,et al.Runs of homozygosity detection and selection signature analysis for local goat breeds in yunnan, China[J].Genes,2024,15(3):313.
doi: 10.3390/genes15030313 |
| 44 |
SHIL,WANGL,LIUJ,et al.Estimation of inbreeding and identification of regions under heavy selection based on runs of homozygosity in a large white pig population[J].J Anim Sci Biotechnol,2020,11(1):46.
doi: 10.1186/s40104-020-00447-0 |
| 45 |
FREITASP H F,WANGY,YANP,et al.Genetic diversity and signatures of selection for thermal stress in cattle and other two bos species adapted to divergent climatic conditions[J].Front Genet,2021,12,604823.
doi: 10.3389/fgene.2021.604823 |
| 46 |
WANGJ.Marker-based estimates of relatedness and inbreeding coefficients: an assessment of current methods[J].J Evol Biol,2014,27(3):518-530.
doi: 10.1111/jeb.12315 |
| 47 |
LIG,TANGJ,HUANGJ,et al.Genome-wide estimates of runs of homozygosity, heterozygosity, and genetic load in two chinese indigenous goat breeds[J].Front Genet,2022,13,774196.
doi: 10.3389/fgene.2022.774196 |
| 48 |
王浩宇,马克岩,李讨讨,等.基于简化基因组测序评估小骨山羊群体遗传多样性和群体结构[J].畜牧兽医学报,2025,56(3):1170-1179.
doi: 10.11843/j.issn.0366-6964.2025.03.018 |
|
WANGH Y,MAK Y,LIT T,et al.Population genetic diversity and population structure analysis of Small-boned goat based on specific-locus amplified fragment sequencing[J].Acta Veterinaria et Zootechnica Sinica,2025,56(3):1170-1179.
doi: 10.11843/j.issn.0366-6964.2025.03.018 |
|
| 49 | JOLLIFFEI T,CADIMAJ.Principal component analysis: a review and recent developments[J].Philo Trans A Math Phys Eng Sci,2016,374(2065):20150202. |
| 50 |
KAPLIP,YANGZ,TELFORDM J.Phylogenetic tree building in the genomic age[J].Nat Rev Genet,2020,21(7):428-444.
doi: 10.1038/s41576-020-0233-0 |
| 51 |
WANGS,DELEONC,SUNW,et al.Alternative splicing of latrophilin-3 controls synapse formation[J].Nature,2024,626(7997):128-135.
doi: 10.1038/s41586-023-06913-9 |
| 52 |
TOYAS,STRUYFS,HUERTAL,et al.A narrative review of chemokine receptors CXCR1 and CXCR2 and their role in acute respiratory distress syndrome[J].Eur Respir Rev,2024,33(173):230172.
doi: 10.1183/16000617.0172-2023 |
| 53 |
HUANGS,XUP,SHEND D,et al.GPCRs steer gi and gs selectivity via TM5-TM6 switches as revealed by structures of serotonin receptors[J].Mol Cell,2022,82(14):2681-2695.e6.
doi: 10.1016/j.molcel.2022.05.031 |
| 54 |
WENX,SHANGP,CHENH,et al.Evolutionary study and structural basis of proton sensing by mus GPR4 and xenopus GPR4[J].Cell,2025,188(3):653-670.e24.
doi: 10.1016/j.cell.2024.12.001 |
| [1] | LIU Sha, YANG Caichun, ZHANG Xiaoyu, CHEN Qiong, LIU Xiong, CHEN Hongbo, ZHOU Huanhuan, SHI Liangyu. Population Genetic Structure and Genome-wide Runs of Homozygosity Analysis in Meihuaxing Pigs Based on 80K SNP Chip [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(8): 3749-3760. |
| [2] | REN Qianzi, ZHANG Baizhong, WANG Zhenqing, WANG Xianglin, GONG Ying, HU Renke, PU Yabin, SU Peng, LI Yefang, MA Yuehui, LI Haobang, JIANG Lin. Genetic Evolutionary Analysis of Wuxue Goat Based on Whole Genome Resequencing [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(8): 3787-3801. |
| [3] | MIAO Junjie, ZHANG Riquan, WU Houyi, YOU Xinming, HUANG Yiwen, HUANG Xiaoying, GUO Zhenyang, LIU Jianlin, XIAO Weihua, GUO Tianhua, CHEN Hao, KANG Dongliu. Genome-Wide SNP Analysis Revealed the Characteristics of Germplasm Resources and Genetic Diversity of Jinggang Black-Palm Geese [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(7): 3199-3209. |
| [4] | WANG Qinqian, GAO Zhendong, LU Ying, MA Ruoshan, DENG Weidong, HE Xiaoming. Research Progress of Whole Genome Resequencing in Chinese Indigenous Cattle [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(5): 2026-2037. |
| [5] | YAO Tingting, LI Hao, YAN Huixuan, CAO Yifan, Cirengluobu , Suolangquji , Nimacangjue , ZHAO Li, Danzengluosang , Silangwangmu , Basangzhuzha , CHEN Ningbo. Genetic Diversity of Mitochondrial Genome and Maternal Origin of 10 Cattle Populations in Tibet Autonomous Region [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(5): 2194-2202. |
| [6] | WANG Haoyu, MA Keyan, LI Taotao, LI Dengpan, ZHAO Qing, MA Youji. Population Genetic Diversity and Population Structure Analysis of Small-boned Goat Based on Specific-Locus Amplified Fragment Sequencing [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(3): 1170-1179. |
| [7] | HU Xin, YOU Wei, JIANG Fugui, CHENG Haijian, SUN Zhigang, SONG Enliang. Analysis of Genetic Diversity and Population Structure of Simmental Cattle Based on Whole Genome Resequencing [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(3): 1189-1202. |
| [8] | LI Yaxuan, SHAO Changliang, GAO Haoran, WU Jinshan, XU Mengqi, WANG Yipeng, LIU Haojun, SU Jingyu, CHEN Junhua, LI Mengxin, MA Yingjie, SHAN Wenjuan. Genetic Diversity and Structure Analysis of the Equus hemionus hemionusin Kalamaili [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(10): 4973-4987. |
| [9] | Siyu LIU, Man ZHANG, Yan ZHANG, Zhitong WEI, Xinglei QI, Tengyun GAO, Xian LIU, Dong LIANG, Tong FU. Evaluation of the Conservation Effect in Nanyang Cattle Based on Resequencing Data [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(9): 3876-3886. |
| [10] | Hongyan HUANG, Liyun ZHANG, Zhirong HUANG, Zhongping WU, Xumeng ZHANG, Hongjia OUYANG, Junpeng CHEN, Zhenping LIN, Yunbo TIAN, Xiujin LI, Yunmao HUANG. The Study on Population Genetic Diversity and Genome-wide Association Study of Body Weight and Size Traits for Lion-head Geese [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(9): 3914-3924. |
| [11] | Tao ZHANG, Jiaqi LI, Lei XU, Dan WANG, Menghua ZHANG, Tao ZHANG, Mengjie YAN, Weitao WANG, Shoumin FAN, Xixia HUANG. Detection and Population Structure Analysis of Genomic Structural Variation in Xinjiang Brown Cattle Based on Whole Genome Resequencing Data [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(8): 3427-3435. |
| [12] | SONG Kelin, YAN Zunqiang, WANG Pengfei, CHENG Wenhao, LI Jie, BAI Yaqin, SUN Guohu, GUN Shuangbao. Analysis on Genetic Diversity and Genetic Structure Based on SNP Chips of Huixian Qingni Black Pig [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(3): 995-1006. |
| [13] | REN Yuwei, CHEN Xing, LIN Yanning, HUANG Xiaoxian, HONG Lingling, WANG Feng, SUN Ruiping, ZHANG Yan, LIU Hailong, ZHENG Xinli, CHAO Zhe. Investigating the Influencing Factors of Egg Laying Performance in Wenchang Chickens Based on Whole Genome Resequencing [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(2): 502-514. |
| [14] | CHENG Xinyan, WANG Shiyuan, JI Yebiao, HUANG Sixiu, YANG Jie, MENG Fanming, ZHANG Mao, CAI Gengyuan, LIU Langqing. Evaluation of the Genetic Structure of Conservation Populations of Four Major Local Pig Breeds in Guangdong Province Based on a 50K SNP Chip [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(12): 5464-5477. |
| [15] | XU Kuowei, LI Zhuohui, LENG Tangjian, XIONG Bao, ZHOU Jielong, GUO Panjiang, WANG Yu, CHEN Fenfen. Analysis of Population Genetic Diversity and Population Genetic Structure of Conservation Population in Ninglang Plateau Chickens Based on Whole-genome Resequencing SNP [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(12): 5498-5510. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||