





Acta Veterinaria et Zootechnica Sinica ›› 2024, Vol. 55 ›› Issue (12): 5498-5510.doi: 10.11843/j.issn.0366-6964.2024.12.016
• Animal Genetics and Breeding • Previous Articles Next Articles
XU Kuowei1( ), LI Zhuohui2(
), LI Zhuohui2( ), LENG Tangjian1, XIONG Bao3, ZHOU Jielong1, GUO Panjiang1, WANG Yu2,*(
), LENG Tangjian1, XIONG Bao3, ZHOU Jielong1, GUO Panjiang1, WANG Yu2,*( ), CHEN Fenfen1,*(
), CHEN Fenfen1,*( )
)
Received:2024-05-22
Online:2024-12-23
Published:2024-12-27
Contact:
WANG Yu, CHEN Fenfen
E-mail:xukuowei@foxmail.com;lizhuohui2021@nwafu.edu.cn;wang_yu@nwsuaf.edu.cn;ffchen03@sina.com
CLC Number:
XU Kuowei, LI Zhuohui, LENG Tangjian, XIONG Bao, ZHOU Jielong, GUO Panjiang, WANG Yu, CHEN Fenfen. Analysis of Population Genetic Diversity and Population Genetic Structure of Conservation Population in Ninglang Plateau Chickens Based on Whole-genome Resequencing SNP[J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(12): 5498-5510.
Table 1
Population genetic diversity index"
| 品种 Breed | 观测杂合度(Ho) Observed heterozygosity | 期望杂合度(He) Expected heterozygosity | 多态标记比例(PN) Polymorphic marker ratio | 核苷酸多态性(Pi) Nucleotide diversity | 次等位基因频率(Maf) Minor allele frequency |
| 大围山微型鸡Daweishan | 0.232 | 0.229 | 0.819 0 | 4.04×10-3 | 0.164 |
| 独龙鸡Dulong | 0.239 | 0.225 | 0.736 7 | 4.06×10-3 | 0.162 |
| 尼西鸡Nixi | 0.231 | 0.219 | 0.730 6 | 3.83×10-3 | 0.158 |
| 宁蒗高原鸡Ninglang | 0.212 | 0.221 | 0.868 3 | 3.94×10-3 | 0.159 |

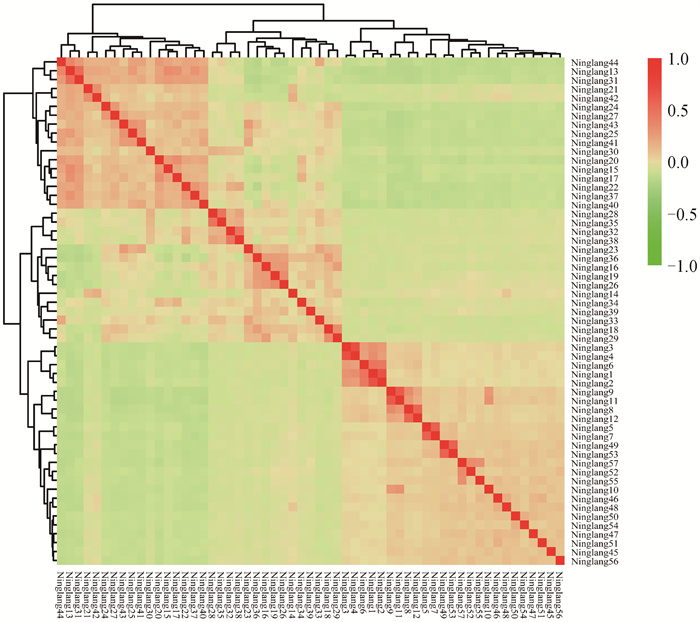
Fig. 7
Visualization results of IBS genetic distance matrix in Ninglang Plateau chickens population The horizontal and vertical coordinates represent the Ninglang Plateau chickens individuals in this study. Each small square represents the genetic distance value between two individuals. The closer the color is to red, the greater the genetic distance between the two individuals, and vice versa"


Fig. 8
Results of G matrix analysis of genetic relationships among Ninglang Plateau chickens populations The horizontal and vertical coordinates are 57 Ninglang Plateau chickens individuals. Each small square represents the genetic relationship between two individuals. The closer the color is to red, the closer the genetic relationship between the two individuals, and vice versa"
| 1 | [1]TAO W K, ANIWARL, ZULIPICARA, 等. Analysis of genetic diversity and population structure of Tarim and Junggar Bactrian camels based on simplified GBS genome sequencing[J]. Animals (Basel), 2023, 13 (14): 2349. |
| 2 |
CHEN J , ZHANG L L , GAO L T , et al. Population structure and genetic diversity of Yunling cattle determined by whole-genome resequencing[J]. Genes (Basel), 2023, 14 (12): 2141.
doi: 10.3390/genes14122141 |
| 3 |
SUN Q X , WANG M G , LU T , et al. Differentiated adaptative genetic architecture and language-related demographical history in South China inferred from 619 genomes from 56 populations[J]. BMC Biol, 2024, 22 (1): 55.
doi: 10.1186/s12915-024-01854-9 |
| 4 |
DEMENTIEVA N V , SHCHERBAKOV Y S , TYSHCHENKO V I , et al. Comparative analysis of molecular RFLP and SNP markers in assessing and understanding the genetic diversity of various chicken breeds[J]. Genes (Basel), 2022, 13 (10): 1876.
doi: 10.3390/genes13101876 |
| 5 |
HUO J L , WU G S , CHEN T , et al. Genetic diversity of local Yunnan chicken breeds and their relationships with Red Junglefowl[J]. Genet Mol Res, 2014, 13 (2): 3371- 3383.
doi: 10.4238/2014.April.29.16 |
| 6 |
ISLAM M A , OSMAN S A M , NISHIBORI M . Genetic diversity of Bangladeshi native chickens based on complete sequence of mitochondrial DNA D-loop region[J]. Br Poult Sci, 2019, 60 (6): 628- 637.
doi: 10.1080/00071668.2019.1655708 |
| 7 |
WANG M S , THAKUR M , PENG M S , et al. 863 genomes reveal the origin and domestication of chicken[J]. Cell Res, 2020, 30 (8): 693- 701.
doi: 10.1038/s41422-020-0349-y |
| 8 |
ISHENGOMA D S , MANDARA C I , MADEBE R A , et al. Microsatellites reveal high polymorphism and high potential for use in anti-malarial efficacy studies in areas with different transmission intensities in mainland Tanzania[J]. Malar J, 2024, 23 (1): 79.
doi: 10.1186/s12936-024-04901-6 |
| 9 |
NEALE D B , KREMER A . Forest tree genomics: growing resources and applications[J]. Nat Rev Genet, 2011, 12 (2): 111- 122.
doi: 10.1038/nrg2931 |
| 10 |
GU J J , LI S , ZHU B , et al. Genetic variation and domestication of horses revealed by 10 chromosome-level genomes and whole-genome resequencing[J]. Mol Ecol Resour, 2023, 23 (7): 1656- 1672.
doi: 10.1111/1755-0998.13818 |
| 11 |
TEREFE E , BELAY G , TIJJANI A , et al. Whole genome resequencing reveals genetic diversity and selection signatures of Ethiopian indigenous cattle adapted to local environments[J]. Diversity, 2023, 15 (4): 540.
doi: 10.3390/d15040540 |
| 12 | XIONG J K , BAO J J , HU W P , et al. Whole-genome resequencing reveals genetic diversity and selection characteristics of dairy goat[J]. Front Genet, 2022, 13, 1044017. |
| 13 | WANG F F , ZHA Z L , HE Y Z , et al. Genome-wide re-sequencing data reveals the population structure and selection signatures of Tunchang pigs in China[J]. Animals (Basel), 2023, 13 (11): 1835. |
| 14 |
CHO Y , KIM J Y , KIM N . Comparative genomics and selection analysis of Yeonsan Ogye black chicken with whole-genome sequencing[J]. Genomics, 2022, 114 (2): 110298.
doi: 10.1016/j.ygeno.2022.110298 |
| 15 |
SUN J L , CHEN T , ZHU M , et al. Whole-genome sequencing revealed genetic diversity and selection of Guangxi indigenous chickens[J]. PLoS One, 2022, 17 (3): e0250392.
doi: 10.1371/journal.pone.0250392 |
| 16 |
SHI S R , SHAO D , YANG L Y , et al. Whole genome analyses reveal novel genes associated with chicken adaptation to tropical and frigid environments[J]. J Adv Res, 2023, 47, 13- 25.
doi: 10.1016/j.jare.2022.07.005 |
| 17 |
RACHMAN M P , BAMIDELE O , DESSIE T , et al. Genomic analysis of Nigerian indigenous chickens reveals their genetic diversity and adaptation to heat-stress[J]. Sci Rep, 2024, 14 (1): 2209.
doi: 10.1038/s41598-024-52569-4 |
| 18 |
WU S W , DOU T F , WANG K , et al. Artificial selection footprints in indigenous and commercial chicken genomes[J]. BMC Genomics, 2024, 25 (1): 428.
doi: 10.1186/s12864-024-10291-5 |
| 19 |
WILKINSON S , WIENER P , TEVERSON D , et al. Characterization of the genetic diversity, structure and admixture of British chicken breeds[J]. Anim Genet, 2012, 43 (5): 552- 563.
doi: 10.1111/j.1365-2052.2011.02296.x |
| 20 |
ZHU W Q , LI H F , WANG J Y , et al. Molecular genetic diversity and maternal origin of Chinese black-bone chicken breeds[J]. Genet Mol Res, 2014, 13 (2): 3275- 3282.
doi: 10.4238/2014.April.29.5 |
| 21 |
PENG M S , HAN J L , ZHANG Y P . Missing puzzle piece for the origins of domestic chickens[J]. Proc Natl Acad Sci U S A, 2022, 119 (44): e2210996119.
doi: 10.1073/pnas.2210996119 |
| 22 | 王欣, 罗成峰, 陶清海, 等. 拉伯高脚鸡线粒体DNA D-loop序列变异与起源分化研究[J]. 中国家禽, 2016, 38 (11): 14- 18. |
| WANG X , LUO C F , TAO Q H , et al. Sequence variation of mtDNA D-loop and origin of Labai high-leg chicken[J]. China Poultry, 2016, 38 (11): 14- 18. | |
| 23 | 许文坤, 刘艺端, 孙利民, 等. 云南省地方家禽遗传资源介绍[J]. 云南农业, 2021, (4): 87- 89. |
| XU W K , LIU Y D , SUN L M , et al. Introduction to local poultry genetic resources in Yunnan Province[J]. Yunnan Agriculture, 2021, (4): 87- 89. | |
| 24 |
邓绍志. 拉伯高脚鸡简介与展望[J]. 中国畜禽种业, 2017, 13 (3): 133- 135.
doi: 10.3969/j.issn.1673-4556.2017.03.117 |
|
DENG S Z . Introduction and prospects of Labai high-leg chicken[J]. The Chinese Livestock and Poultry Breeding, 2017, 13 (3): 133- 135.
doi: 10.3969/j.issn.1673-4556.2017.03.117 |
|
| 25 |
LI H , DURBIN R . Fast and accurate long-read alignment with Burrows-Wheeler transform[J]. Bioinformatics, 2010, 26 (5): 589- 595.
doi: 10.1093/bioinformatics/btp698 |
| 26 |
LI H , HANDSAKER B , WYSOKER A , et al. The sequence alignment/map format and SAMtools[J]. Bioinformatics, 2009, 25 (16): 2078- 2079.
doi: 10.1093/bioinformatics/btp352 |
| 27 |
MCKENNA A , HANNA M , BANKS E , et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data[J]. Genome Res, 2010, 20 (9): 1297- 1303.
doi: 10.1101/gr.107524.110 |
| 28 |
WANG K , LI M , HAKONARSON H . ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data[J]. Nucleic Acids Res, 2010, 38 (16): e164.
doi: 10.1093/nar/gkq603 |
| 29 |
王婷, 张元庆, 闫益波, 等. "特藏寒羊"群体遗传结构分析与选择信号的对比分析[J]. 畜牧兽医学报, 2024, 55 (7): 2913- 2926.
doi: 10.11843/j.issn.0366-6964.2024.07.012 |
|
WANG T , ZHANG Y Q , YAN Y B , et al. The genetic structure analysis and the comparative analysis of selection signals in 'Tezanghan' sheep[J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55 (7): 2913- 2926.
doi: 10.11843/j.issn.0366-6964.2024.07.012 |
|
| 30 |
PURCELL S , NEALE B , TODD-BROWN K , et al. PLINK: a tool set for whole-genome association and population-based linkage analyses[J]. Am J Hum Genet, 2007, 81 (3): 559- 575.
doi: 10.1086/519795 |
| 31 | 张小键. 五个湖北地方鸡品种遗传多样性评估及重要经济性状选择信号鉴定[D]. 武汉: 华中农业大学, 2022. |
| ZHANG X J. Evaluation of genetic diversity of five Hubei native chicken breeds and identification of selective signals for important economic traits[D]. Wuhan: Huazhong Agricultural University, 2022. (in Chinese) | |
| 32 |
DANECEK P , AUTON A , ABECASIS G , et al. The variant call format and VCFtools[J]. Bioinformatics, 2011, 27 (15): 2156- 2158.
doi: 10.1093/bioinformatics/btr330 |
| 33 |
ZHANG C , DONG S S , XU J Y , et al. PopLDdecay: a fast and effective tool for linkage disequilibrium decay analysis based on variant call format files[J]. Bioinformatics, 2019, 35 (10): 1786- 1788.
doi: 10.1093/bioinformatics/bty875 |
| 34 |
ZHANG M M , HAN W , TANG H , et al. Genomic diversity dynamics in conserved chicken populations are revealed by genome-wide SNPs[J]. BMC Genomics, 2018, 19 (1): 598.
doi: 10.1186/s12864-018-4973-6 |
| 35 |
LEE T H , GUO H , WANG X Y , et al. SNPhylo: a pipeline to construct a phylogenetic tree from huge SNP data[J]. BMC Genomics, 2014, 15, 162.
doi: 10.1186/1471-2164-15-162 |
| 36 |
XIE J M , CHEN Y R , CAI G J , et al. Tree Visualization By One Table (tvBOT): a web application for visualizing, modifying and annotating phylogenetic trees[J]. Nucleic Acids Res, 2023, 51 (W1): W587- W592.
doi: 10.1093/nar/gkad359 |
| 37 |
ALEXANDER D H , NOVEMBRE J , LANGE K . Fast model-based estimation of ancestry in unrelated individuals[J]. Genome Res, 2009, 19 (9): 1655- 1664.
doi: 10.1101/gr.094052.109 |
| 38 | MAGLO K N , MERSHA T B , MARTIN L J . Population genomics and the statistical values of race: an interdisciplinary perspective on the biological classification of human populations and implications for clinical genetic epidemiological research[J]. Front Genet, 2016, 7, 22. |
| 39 |
YANG J , LEE S H , GODDARD M E , et al. GCTA: a tool for genome-wide complex trait analysis[J]. Am J Hum Genet, 2011, 88 (1): 76- 82.
doi: 10.1016/j.ajhg.2010.11.011 |
| 40 |
MCQUILLAN R , LEUTENEGGER A L , ABDEL-RAHMAN R , et al. Runs of homozygosity in European populations[J]. Am J Hum Genet, 2008, 83 (3): 359- 372.
doi: 10.1016/j.ajhg.2008.08.007 |
| 41 |
宋科林, 闫尊强, 王鹏飞, 等. 基于SNP芯片分析徽县青泥黑猪遗传多样性和遗传结构[J]. 畜牧兽医学报, 2024, 55 (3): 995- 1006.
doi: 10.11843/j.issn.0366-6964.2024.03.013 |
|
SONG K L , YAN Z Q , WANG P F , et al. Analysis on genetic diversity and genetic structure based on SNP chips of Huixian Qingni Black pig[J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55 (3): 995- 1006.
doi: 10.11843/j.issn.0366-6964.2024.03.013 |
|
| 42 |
SARTIKA T , SAPUTRA F , TAKAHASHI H . Genetic diversity of eight native indonesian chicken breeds on microsatellite markers[J]. HAYATI J Biosci, 2022, 30 (1): 122- 130.
doi: 10.4308/hjb.30.1.122-130 |
| 43 |
TIAN S S , LI W , ZHONG Z Q , et al. Genome-wide re-sequencing data reveals the genetic diversity and population structure of Wenchang chicken in China[J]. Anim Genet, 2023, 54 (3): 328- 337.
doi: 10.1111/age.13293 |
| 44 |
XU D , ZHU W , WU Y H , et al. Whole-genome sequencing revealed genetic diversity, structure and patterns of selection in Guizhou indigenous chickens[J]. BMC Genomics, 2023, 24 (1): 570.
doi: 10.1186/s12864-023-09621-w |
| 45 |
KON T , PEI L Y , ICHIKAWA R , et al. Whole-genome resequencing of large yellow croaker (Larimichthys crocea) reveals the population structure and signatures of environmental adaptation[J]. Sci Rep, 2021, 11 (1): 11235.
doi: 10.1038/s41598-021-90645-1 |
| 46 |
SCHMIDT T L , JASPER M E , WEEKS A R , et al. Unbiased population heterozygosity estimates from genome-wide sequence data[J]. Methods Ecol Evol, 2021, 12 (10): 1888- 1898.
doi: 10.1111/2041-210X.13659 |
| 47 |
BORTOLUZZI C , CROOIJMANS R P M A , BOSSE M , et al. The effects of recent changes in breeding preferences on maintaining traditional Dutch chicken genomic diversity[J]. Heredity (Edinb), 2018, 121 (6): 564- 578.
doi: 10.1038/s41437-018-0072-3 |
| 48 |
LIU J J , XIAO Y , REN P W , et al. Integrating genomics and transcriptomics to identify candidate genes for high egg production in Wulong geese (Anser cygnoides orientalis)[J]. BMC Genomics, 2023, 24 (1): 481.
doi: 10.1186/s12864-023-09603-y |
| 49 | 肖倩. 浦东白猪种质特性及其保护与利用研究[D]. 上海: 上海交通大学, 2017. |
| XIAO Q. Study on breed characters, conservation and utilization of Pudong White pigs[D]. Shanghai: Shanghai Jiao Tong University, 2017. (in Chinese) | |
| 50 | 李凯航, 赵乐乐, 陆雪林, 等. 基于SNP芯片的浦东鸡保种分析[J]. 中国家禽, 2020, 42 (6): 31- 36. |
| LI K H , ZHAO L L , LU X L , et al. Analysis of conservation effect in Pudong chicken based on SNP chip[J]. China Poultry, 2020, 42 (6): 31- 36. | |
| 51 | 周明芳. 三个不同地理分布丝羽乌骨鸡群体遗传多样性、遗传结构及选择信号分析[D]. 南昌: 江西农业大学, 2022. |
| ZHOU M F. Genetic diversity, genetic structure and selection signal analysis of three different regional Silkies (Gallus gallus domesticus Brisson)[D]. Nanchang: Jiangxi Agricultural University, 2022. (in Chinese) | |
| 52 | 李德娟, 朱迪, 张浩, 等. 基于全基因组SNPs对太行鸡保种群保种效果的评价[J]. 中国畜牧杂志, 2024, 60 (3): 118- 126. |
| LI D J , ZHU D , ZHANG H , et al. Evaluation of the conservation effect of Taihang chicken conservation population based on whole genome SNPs[J]. Chinese Journal of Animal Science, 2024, 60 (3): 118- 126. | |
| 53 |
ZHANG M M , WANG S W , XU R , et al. Managing genomic diversity in conservation programs of Chinese domestic chickens[J]. Genet Sel Evol, 2023, 55 (1): 92.
doi: 10.1186/s12711-023-00866-3 |
| 54 | GAO C Q , DU W P , TIAN K T , et al. Analysis of conservation priorities and runs of homozygosity patterns for Chinese indigenous chicken breeds[J]. Animals (Basel), 2023, 13 (4): 599. |
| 55 | ZHANG J X , NIE C S , LI X H , et al. Genome-wide population genetic analysis of commercial, indigenous, game, and wild chickens using 600K SNP microarray data[J]. Front Genet, 2020, 11, 543294. |
| 56 | TOLONE M , SARDINA M T , CRISCIONE A , et al. High-density single nucleotide polymorphism markers reveal the population structure of 2 local chicken genetic resources[J]. Poult Sci, 2023, 102 (7): 102692. |
| 57 | KARDOS M , ARMSTRONG E E , FITZPATRICK S W , et al. The crucial role of genome-wide genetic variation in conservation[J]. Proc Natl Acad Sci U S A, 2021, 118 (48): e2104642118. |
| [1] | Siyu LIU, Man ZHANG, Yan ZHANG, Zhitong WEI, Xinglei QI, Tengyun GAO, Xian LIU, Dong LIANG, Tong FU. Evaluation of the Conservation Effect in Nanyang Cattle Based on Resequencing Data [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(9): 3876-3886. |
| [2] | Hongyan HUANG, Liyun ZHANG, Zhirong HUANG, Zhongping WU, Xumeng ZHANG, Hongjia OUYANG, Junpeng CHEN, Zhenping LIN, Yunbo TIAN, Xiujin LI, Yunmao HUANG. The Study on Population Genetic Diversity and Genome-wide Association Study of Body Weight and Size Traits for Lion-head Geese [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(9): 3914-3924. |
| [3] | Wei LIU, Jiayi MA, Haoyu GENG, Tian XIE, Sunan MIAO, Zongjie LIAO, Shizhong GENG. Isolation, Identification and Characterization of a Broad Spectrum Salmonella Phage [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(9): 4061-4068. |
| [4] | Tao ZHANG, Jiaqi LI, Lei XU, Dan WANG, Menghua ZHANG, Tao ZHANG, Mengjie YAN, Weitao WANG, Shoumin FAN, Xixia HUANG. Detection and Population Structure Analysis of Genomic Structural Variation in Xinjiang Brown Cattle Based on Whole Genome Resequencing Data [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(8): 3427-3435. |
| [5] | Yifan NIU, Chongyang LI, Baigao YANG, Peipei ZHANG, Hang ZHANG, Xiaoyi FENG, Jianhua CAO, Zhou YU, Youji MA, Xueming ZHAO. Evaluation of the Effect of Different Single Cell Whole Genome Amplification Systems on the Amplification of Bovine Trace Blood DNA [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(8): 3436-3445. |
| [6] | Ting WANG, Yuanqing ZHANG, Yibo YAN, Mingjun SHANGGUAN, Hongyu GUO, Zhiwu WANG. The Genetic Structure Analysis and the Comparative Analysis of Selection Signals in 'Tezanghan' Sheep [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(7): 2913-2926. |
| [7] | TU Yun, ZENG Yanan, ZHANG Zhenghao, HONG Rui, WANG Zhen, WU Ping, ZHOU Zeyang, YE Yiru, DU Yanan, ZUO Fuyuan, ZHANG Gongwei. Genetic Structure and Runs of Homozygosity Analysis of Fuling Buffalo and Southwest Buffalo Breeds [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(5): 1989-1998. |
| [8] | TIAN Rui, XU Sixiang, XIE Feng, LIU Guangjin, WANG Gang, LI Qingxia, DAI Lei, XIE Guoxin, ZHANG Qiongwen, LU Yajing, WANG Guangwen, WANG Jinxiu, ZHANG Wei. Bioinformatics Analysis of the Genome of Clostridium perfringens Isolated from Cattle [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(4): 1707-1715. |
| [9] | REN Yuwei, CHEN Xing, LIN Yanning, HUANG Xiaoxian, HONG Lingling, WANG Feng, SUN Ruiping, ZHANG Yan, LIU Hailong, ZHENG Xinli, CHAO Zhe. Investigating the Influencing Factors of Egg Laying Performance in Wenchang Chickens Based on Whole Genome Resequencing [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(2): 502-514. |
| [10] | CHENG Xinyan, WANG Shiyuan, JI Yebiao, HUANG Sixiu, YANG Jie, MENG Fanming, ZHANG Mao, CAI Gengyuan, LIU Langqing. Evaluation of the Genetic Structure of Conservation Populations of Four Major Local Pig Breeds in Guangdong Province Based on a 50K SNP Chip [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(12): 5464-5477. |
| [11] | GAO Jiaojiao, ZHENG Nan, SHAO Wei, CHEN He, MA Xianlan, ZHAO Yankun. Characterization of Heterogeneous Drug-resistant Escherichia coli and Its Drug-resistant Subpopulations from Milk Sources [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(12): 5813-5824. |
| [12] | Huili LIANG, Yujing XIE, Bowen SI, Guiying WANG, Yunliang JIANG, Guiling CAO. Analysis on Genomic Variation and Population Structure of Large-tailed Han Sheep Based on Whole Genome Resequencing [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(11): 4968-4979. |
| [13] | Zhenni LIU, Jianjun LI, Hai LIAN, Xiaowen LEI, Donghai TAN, Qingyuan ZENG, Di CHENG, Yuling TIAN, Zhiwei KONG, Hualiang XIE, Yunping ZHONG. Population Evolution Analysis of Ganzhou Muscovy Duck Based on Whole Genome Resequencing [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(11): 4992-5002. |
| [14] | Qingzhen SHI, Hongyang XU, Yan ZHANG, Yi ZHANG, Yachun WANG, Jianyong HAN, Li JIANG. Detection and Comparative Analysis of Genomic Genetic Variations in Trace Cells Using Different Methods [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(10): 4311-4324. |
| [15] | YUAN Wei, BI Huan, ZHANG Yudan, ZHANG Yiyu, GU Xiaolong, YANG Hongwen, CHEN Wei. Deciphering Genome-wide Selection Signals Reveals Genetic Differences between Jianbai and Congjiang Xiang Pigs [J]. Acta Veterinaria et Zootechnica Sinica, 2023, 54(9): 3631-3641. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||