





Acta Veterinaria et Zootechnica Sinica ›› 2025, Vol. 56 ›› Issue (7): 3199-3209.doi: 10.11843/j.issn.0366-6964.2025.07.015
• Animal Genetics and Breeding • Previous Articles Next Articles
MIAO Junjie1,2( ), ZHANG Riquan3, WU Houyi3, YOU Xinming3, HUANG Yiwen1, HUANG Xiaoying3, GUO Zhenyang4, LIU Jianlin5, XIAO Weihua6, GUO Tianhua6, CHEN Hao2, KANG Dongliu1,*(
), ZHANG Riquan3, WU Houyi3, YOU Xinming3, HUANG Yiwen1, HUANG Xiaoying3, GUO Zhenyang4, LIU Jianlin5, XIAO Weihua6, GUO Tianhua6, CHEN Hao2, KANG Dongliu1,*( )
)
Received:2024-12-26
Online:2025-07-23
Published:2025-07-25
Contact:
KANG Dongliu
E-mail:1171708961@qq.com;kangdongliu@sina.com
CLC Number:
MIAO Junjie, ZHANG Riquan, WU Houyi, YOU Xinming, HUANG Yiwen, HUANG Xiaoying, GUO Zhenyang, LIU Jianlin, XIAO Weihua, GUO Tianhua, CHEN Hao, KANG Dongliu. Genome-Wide SNP Analysis Revealed the Characteristics of Germplasm Resources and Genetic Diversity of Jinggang Black-Palm Geese[J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(7): 3199-3209.
Table 1
Samples origin"
| 品种 Breed | 品种代号 Breed code | 样本量 Sample size | 原产地 Origin region | 体型 Body type |
| 定安鹅Dingan goose | DAG | 20 | 海南定安 | 中等体型 |
| 丰城灰鹅Fengcheng grey goose | FCG | 20 | 江西丰城 | 中等体型 |
| 井冈黑掌鹅Jinggang black-palm goose | JGG | 81 | 江西吉安 | 大体型 |
| 马岗鹅Magang goose | MGG | 20 | 广东开平 | 中等体型 |
| 狮头鹅Shitou goose | STG | 20 | 广东饶平 | 大体型 |
| 乌鬃鹅Wuzong goose | WZG | 20 | 广东清远 | 小体型 |
| 兴国灰鹅Xingguo grey goose | XGG | 20 | 江西兴国 | 中等体型 |
| 总计Total | 201 |
Table 2
Comparison of genetic diversity between Jinggang black-palm geese and other geese populations"
| 品种 Breed | 缩写 Breed code | 数量 Sample size | CSNPs | Pn | He | Ho | F | DST | ROH |
| 定安鹅Dingan goose | DAG | 20 | 3 967 550 | 0.93 | 0.30 | 0.27 | 0.16±0.04 | 0.260±0.012 | 5.411±3.194 |
| 丰城灰鹅 Fengcheng grey goose | FCG | 20 | 3 956 616 | 0.93 | 0.30 | 0.27 | 0.16±0.05 | 0.259±0.004 | 4.583±1.019 |
| 井冈黑掌鹅 Jinggang black-palm goose | JGG | 20 | 3 414 168 | 0.83 | 0.26 | 0.23 | 0.33±0.09 | 0.224±0.022 | 12.874±8.273 |
| 马岗鹅Magang goose | MGG | 20 | 3 799 547 | 0.91 | 0.29 | 0.26 | 0.22±0.11 | 0.252±0.006 | 6.791±1.506 |
| 狮头鹅Shitou goose | STG | 20 | 3 653 824 | 0.86 | 0.27 | 0.25 | 0.24±0.09 | 0.237±0.005 | 9.013±2.019 |
| 乌鬃鹅Wuzong goose | WZG | 20 | 3 550 844 | 0.79 | 0.26 | 0.26 | 0.22±0.03 | 0.216±0.014 | 13.298±2.942 |
| 兴国灰鹅 Xingguo grey goose | XGG | 20 | 3 931 859 | 0.94 | 0.30 | 0.29 | 0.10±0.05 | 0.252±0.007 | 4.909±1.148 |
| 总计Total | 140 |

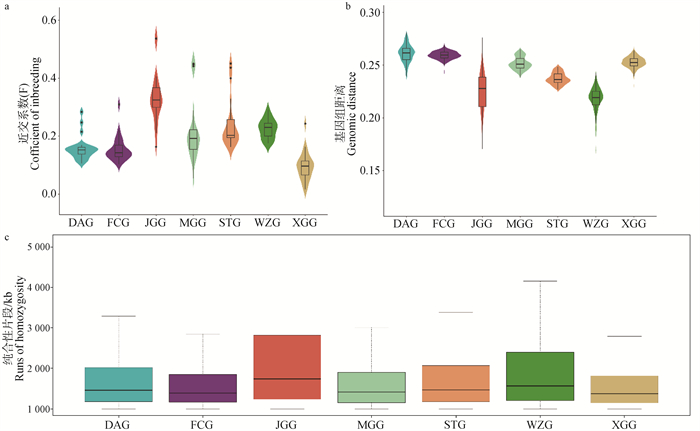
Fig. 7
Comparison of genetic diversity among 7 Chinese domestic geese breeds a. The inbreeding coefficient F of each breed, where a higher value of F represents more frequent inbreeding within the population; b. The level of intra-population genetic distance DST for each breed, with smaller DST representing closer relatedness of individuals within the population; c. Distribution of ROH at the level of homozygous fragments for each breed, with higher and longer ROH indicating lower genetic diversity of the population"
| 1 | EDA M , ITAHASHI Y , KIKUCHI H , et al. Multiple lines of evidence of early goose domestication in a 7, 000-y-old rice cultivation village in the lower Yangtze River, China[J]. Proc Natl Acad Sci U S A, 2022, 119 (12): e2117064119. |
| 2 | REN T , LIANG S , ZHAO A , et al. Analysis of the complete mitochondrial genome of the Zhedong White goose and characterization of NUMTs: reveal domestication history of goose in China and Euro[J]. Gene, 2016, 577 (1): 75- 81. |
| 3 | LI H , ZHU W , CHEN K , et al. Two maternal origins of Chinese domestic goose[J]. Poult Sci, 2011, 90 (12): 2705- 2710. |
| 4 | 国家畜禽遗传资源委员会. 中国畜禽遗传资源志[M]. 北京: 中国农业出版社, 2011. |
| National Commission for Livestock and Poultry Genetic Resources . China Livestock and Poultry Genetic Resources Annals[M]. Beijing: China Agriculture Press, 2011. | |
| 5 | CHO Y , KIM J Y , KIM N . Comparative genomics and selection analysis of Yeonsan Ogye black chicken with whole-genome sequencing[J]. Genomics, 2022, 114 (2): 110298. |
| 6 | CHEN H , LUO K , WANG C , et al. Genomic characteristics and selection signals of Zhongshan ducks[J]. Animal, 2023, 17 (5): 100797. |
| 7 | ZHENG S , OUYANG J , LIU S , et al. Genomic signatures reveal selection in Lingxian white goose[J]. Poult Sci, 2023, 102 (1): 102269. |
| 8 | CAI Z , SARUP P , OSTERSTEN T , et al. Genomic diversity revealed by whole-genome sequencing in three Danish commercial pig breeds[J]. J Anim Sci, 2020, 98 (7): skaa229. |
| 9 | BHATI M , KADRI N K , CRYSNANTO D , et al. Assessing genomic diversity and signatures of selection in Original Braunvieh cattle using whole-genome sequencing data[J]. BMC Genomics, 2020, 21 (1): 1- 14. |
| 10 | BOLGER A M , LOHSE M , USADEL B . Trimmomatic: a flexible trimmer for Illumina sequence data[J]. Bioinformatics, 2014, 30 (15): 2114- 2120. |
| 11 | LI H , DURBIN R . Fast and accurate short read alignment with Burrows-Wheeler transform[J]. Bioinformatics, 2009, 25 (14): 1754- 1760. |
| 12 | LI H , HANDSHAKER B , WYSOKER A , et al. The sequence alignment/map format and SAMtools[J]. Bioinformatics, 2009, 25 (16): 2078- 2079. |
| 13 | MCKENNA A , HANNA M , BANKS E , et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data[J]. Genome Res, 2010, 20 (9): 1297- 1303. |
| 14 | CINGOLANI P , PLATTS A , WANG L L , et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3[J]. Fly, 2012, 6 (2): 80- 92. |
| 15 | CHANG C C , CHOW C C , TELLIER L C , et al. Second-generation PLINK: rising to the challenge of larger and richer datasets[J]. Gigascience, 2015, 4 (1): s13742-015-0047-8. |
| 16 | LETUNIC I , BORK P . Interactive Tree of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation[J]. Nucleic Acids Res, 2021, 49 (W1): W293- W296. |
| 17 | YANG J , LEE S H , GODDARD M E , et al. GCTA: a tool for genome-wide complex trait analysis[J]. Am J Hum Genet, 2011, 88 (1): 76- 82. |
| 18 | ALEXANDER D H , NOVEMBER J , LANGE K . Fast model-based estimation of ancestry in unrelated individuals[J]. Genome Res, 2009, 19 (9): 1655- 1664. |
| 19 | ZHONG Z , WANG Z , XIE X , et al. Evaluation of the genetic diversity, population structure and selection signatures of three native Chinese pig populations[J]. Animals, 2023, 13 (12): 2010. |
| 20 | CHEN N , XIA X , HANIF Q , et al. Global genetic diversity, introgression, and evolutionary adaptation of indicine cattle revealed by whole genome sequencing[J]. Nat Commun, 2023, 14 (1): 7803. |
| 21 |
胡鑫, 游伟, 姜富贵, 等. 基于全基因组重测序分析西门塔尔牛遗传多样性与群体结构[J]. 畜牧兽医学报, 2025, 56 (3): 1189- 1202.
doi: 10.11843/j.issn.0366-6964.2025.03.020 |
|
HU X , YOU W , JIANG F G , et al. Analysis of genetic diversity and population structure of Simmental cattle based on whole genome resequencing[J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56 (3): 1189- 1202.
doi: 10.11843/j.issn.0366-6964.2025.03.020 |
|
| 22 | CENDRON F , MASTRANGELO S , TOLONE M , et al. Genome-wide analysis reveals the patterns of genetic diversity and population structure of 8 Italian local chicken breeds[J]. Poult Sci, 2021, 100 (2): 441- 451. |
| 23 | SHI S , SHAO D , YANG L , et al. Whole genome analyses reveal novel genes associated with chicken adaptation to tropical and frigid environments[J]. J Adv Res, 2023, 47, 13- 25. |
| 24 | SUN J , CHEN T , ZHU M , et al. Whole-genome sequencing revealed genetic diversity and selection of Guangxi indigenous chickens[J]. PLoS One, 2022, 17 (3): e0250392. |
| 25 | LIN R , LI J , YANG Y , et al. Genome-wide population structure analysis and genetic diversity detection of four Chinese indigenous duck breeds from Fujian province[J]. Animals, 2022, 12 (17): 2302. |
| 26 | LI L , QUAN J , GAO C , et al. Whole-genome resequencing to unveil genetic characteristics and selection signatures of specific pathogen-free ducks[J]. Poult Sci, 2023, 102 (7): 102748. |
| 27 | ZHAO Q , LIN Z , CHEN J , et al. Chromosome-level genome assembly of goose provides insight into the adaptation and growth of local goose breeds[J]. Gigascience, 2023, 12, giad003. |
| 28 | GAO G , ZHANG K , HUANG P , et al. Identification of SNPs associated with goose meat quality traits using a genome-wide association study approach[J]. Animals, 2023, 13 (13): 2089. |
| 29 | ZENG N , LIU G , WEN S , et al. New bird records and bird diversity of Poyang Lake national nature reserve, Jiangxi Province, China[J]. Pak J Zool, 2018, 50 (4): 1285- 1291. |
| 30 | GAO G , ZHANG H , NI J , et al. Insights into genetic diversity and phenotypic variations in domestic geese through comprehensive population and pan-genome analysis[J]. J Anim Sci Biotechnol, 2023, 14 (1): 150. |
| 31 | YAO Y , OCHOA A . Limitations of principal components in quantitative genetic association models for human studies[J]. Elife, 2023, 12, e79238. |
| 32 | RODRÍGUEZ-RAMILO S T , GARCÍA-CORTÉS L A , DE CARA M Á R . Artificial selection with traditional or genomic relationships: consequences in coancestry and genetic diversity[J]. Front Genet, 2015, 6, 127. |
| 33 | GUO G , PAN B , YI X , et al. Genetic diversity between local landraces and current breeding lines of pepper in China[J]. Sci Rep, 2023, 13 (1): 4058. |
| 34 | DEMENTIEVA N V , MITROFANOVA O V , DYSIN A P , et al. Assessing the effects of rare alleles and linkage disequilibrium on estimates of genetic diversity in the chicken populations[J]. Animal, 2021, 15 (3): 100171. |
| 35 | HAQUE M A , JUNG J H , CHOO H J , et al. Pedigree analysis of Korean native chickens: unraveling inbreeding and genetic diversity[J]. Poult Sci, 2024, 103 (10): 104071. |
| 36 | HOBAN S , BRUFORD M W , DA SILVA J M , et al. Genetic diversity goals and targets have improved, but remain insufficient for clear implementation of the post-2020 global biodiversity framework[J]. Conserv Genet, 2023, 24 (2): 181- 191. |
| 37 | KARDOS M , ARMSTRONG E E , FITZPATRICK S W , et al. The crucial role of genome-wide genetic variation in conservation[J]. Proc Natl Acad Sci U S A, 2021, 118 (48): e2104642118. |
| 38 | ADHIKARI S , KUMARI J , JACOB S R , et al. Landraces-potential treasure for sustainable wheat improvement[J]. Genet Resour Crop Evol, 2022, 69 (2): 499- 523. |
| [1] | WANG Qinqian, GAO Zhendong, LU Ying, MA Ruoshan, DENG Weidong, HE Xiaoming. Research Progress of Whole Genome Resequencing in Chinese Indigenous Cattle [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(5): 2026-2037. |
| [2] | YAO Tingting, LI Hao, YAN Huixuan, CAO Yifan, Cirengluobu , Suolangquji , Nimacangjue , ZHAO Li, Danzengluosang , Silangwangmu , Basangzhuzha , CHEN Ningbo. Genetic Diversity of Mitochondrial Genome and Maternal Origin of 10 Cattle Populations in Tibet Autonomous Region [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(5): 2194-2202. |
| [3] | WANG Haoyu, MA Keyan, LI Taotao, LI Dengpan, ZHAO Qing, MA Youji. Population Genetic Diversity and Population Structure Analysis of Small-boned Goat Based on Specific-Locus Amplified Fragment Sequencing [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(3): 1170-1179. |
| [4] | HU Xin, YOU Wei, JIANG Fugui, CHENG Haijian, SUN Zhigang, SONG Enliang. Analysis of Genetic Diversity and Population Structure of Simmental Cattle Based on Whole Genome Resequencing [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(3): 1189-1202. |
| [5] | Siyu LIU, Man ZHANG, Yan ZHANG, Zhitong WEI, Xinglei QI, Tengyun GAO, Xian LIU, Dong LIANG, Tong FU. Evaluation of the Conservation Effect in Nanyang Cattle Based on Resequencing Data [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(9): 3876-3886. |
| [6] | Hongyan HUANG, Liyun ZHANG, Zhirong HUANG, Zhongping WU, Xumeng ZHANG, Hongjia OUYANG, Junpeng CHEN, Zhenping LIN, Yunbo TIAN, Xiujin LI, Yunmao HUANG. The Study on Population Genetic Diversity and Genome-wide Association Study of Body Weight and Size Traits for Lion-head Geese [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(9): 3914-3924. |
| [7] | SONG Kelin, YAN Zunqiang, WANG Pengfei, CHENG Wenhao, LI Jie, BAI Yaqin, SUN Guohu, GUN Shuangbao. Analysis on Genetic Diversity and Genetic Structure Based on SNP Chips of Huixian Qingni Black Pig [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(3): 995-1006. |
| [8] | CHENG Xinyan, WANG Shiyuan, JI Yebiao, HUANG Sixiu, YANG Jie, MENG Fanming, ZHANG Mao, CAI Gengyuan, LIU Langqing. Evaluation of the Genetic Structure of Conservation Populations of Four Major Local Pig Breeds in Guangdong Province Based on a 50K SNP Chip [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(12): 5464-5477. |
| [9] | XU Kuowei, LI Zhuohui, LENG Tangjian, XIONG Bao, ZHOU Jielong, GUO Panjiang, WANG Yu, CHEN Fenfen. Analysis of Population Genetic Diversity and Population Genetic Structure of Conservation Population in Ninglang Plateau Chickens Based on Whole-genome Resequencing SNP [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(12): 5498-5510. |
| [10] | Xiangzhen GOU, Junxiang YANG, Zihui ZHAO, Lingxia FENG, Wanhui CHEN, Yujie LI, Zhongyu ZHANG, Keyan MA, Dongping JIANG, Rong CHANG, Yazhou WEN, Ke WANG, Youji MA. Evaluation of the Population Structure of the Ziwuling Black Goat Based on Super-GBS [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(10): 4334-4345. |
| [11] | YANG Qing, GONG Jing, ZHAO Xueyan, ZHU Xiaodong, GENG Liying, ZHANG Chuansheng, WANG Jiying. Comparison of Array and Resequencing in Pig Genetic Structure Studies [J]. Acta Veterinaria et Zootechnica Sinica, 2023, 54(7): 2772-2782. |
| [12] | ZHANG Renbao, ZHOU Donghui, ZHOU Lisheng, GAO Xiaoxiao, LIU Nan, HE Jianning. Analysis of Genetic Structure of Conservation Population in Jining Gray Goats Based on 70 K SNP Chip [J]. Acta Veterinaria et Zootechnica Sinica, 2023, 54(7): 2836-2847. |
| [13] | ZHAO Zhenjian, WANG Shujie, CHEN Dong, JI Xiang, SHEN Qi, YU Yang, CUI Shengdi, WANG Junge, CHEN Ziyang, TANG Guoqing. Population Structure and Genetic Diversity Analysis of Neijiang Pigs Based on Low-coverage Whole Genome Sequencing [J]. Acta Veterinaria et Zootechnica Sinica, 2023, 54(6): 2297-2307. |
| [14] | TAO Xuan, YANG Xuemei, LIANG Yan, LIU Yihui, WANG Yong, KONG Fanjing, LEI Yunfeng, YANG Yuekui, WANG Yan, AN Rui, YANG Kun, Lü Xuebin, HE Zhiping, GU Yiren. Analysis of Genetic Structure of Conservation Population in Yacha Pig Based on SNP Chip [J]. Acta Veterinaria et Zootechnica Sinica, 2023, 54(6): 2308-2319. |
| [15] | LIU Shuang, HE Lixia, MA Jun, FENG Xue, YANG Mengli, WANG Shuzhe, YANG Runjun, FANG Xibi, XIAN Hailong, WANG Yongkang, ZHANG Lupei, MA Yun. Analysis on Genetic Background and Body Size Indexes and Beef-purpose Index of Guyuan Cattle [J]. Acta Veterinaria et Zootechnica Sinica, 2023, 54(6): 2376-2388. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||