





Acta Veterinaria et Zootechnica Sinica ›› 2025, Vol. 56 ›› Issue (10): 4973-4987.doi: 10.11843/j.issn.0366-6964.2025.10.018
• Animal Genetics and Breeding • Previous Articles Next Articles
LI Yaxuan1( ), SHAO Changliang2, GAO Haoran1, WU Jinshan1, XU Mengqi1, WANG Yipeng1, LIU Haojun1, SU Jingyu1, CHEN Junhua1, LI Mengxin1, MA Yingjie1,*(
), SHAO Changliang2, GAO Haoran1, WU Jinshan1, XU Mengqi1, WANG Yipeng1, LIU Haojun1, SU Jingyu1, CHEN Junhua1, LI Mengxin1, MA Yingjie1,*( ), SHAN Wenjuan1,*(
), SHAN Wenjuan1,*( )
)
Received:2025-03-11
Online:2025-10-23
Published:2025-11-01
Contact:
MA Yingjie, SHAN Wenjuan
E-mail:107552201006@stu.xju.edu.cn;mayingjie@xju.edu.cn;swj@xju.edu.cn
CLC Number:
LI Yaxuan, SHAO Changliang, GAO Haoran, WU Jinshan, XU Mengqi, WANG Yipeng, LIU Haojun, SU Jingyu, CHEN Junhua, LI Mengxin, MA Yingjie, SHAN Wenjuan. Genetic Diversity and Structure Analysis of the Equus hemionus hemionus in Kalamaili[J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(10): 4973-4987.
Table 1
Primers information of E. h. hemionus microsatellite loci"
| 位点 Locus | 荧光 Dye | 引物序列(5'→3') Primers sequence | 等位基因大小/bp Allele range | 退火温度/℃ Annealing temperature |
| AHT040 | HEX | GCAAGTTCAGCACCTCCCT TTTATGACACCTGCTGAGAACG | 230 | 58 |
| COR053 | HEX | AATTGACTGTGGAAGCCTTG GGCTGAGGAGTAAGCTGAAAG | 173~197 | 55 |
| AHT021 | TAMRA | TCCAAGTTGCTGAATGGATC ACGGCCTGATTCTCTCTTTG | 199~215 | 58 |
| HMS02 | FAM | ACGGTGGCAACTGCCAAGGAAG CTTGCAGTCGAATGTGTATTAAATG | 218~236 | 63 |
| 1CA41 | TAMRA | CTGGTCAGGCCTATTACCCA AGATATTGGGGGCGGAAG | 245 | 58 |
| 1CA25 | HEX | TCCAATTTTCCCCAATGGTA CTGCATTTTGACAATGGTGG | 206 | 58 |
| 1CA43 | FAM | ATGGCATGATTTGCTTCTCC TGGAAACAACCTAAATGTCCA | 122 | 58 |
| COR082 | HEX | GCTTTTGTTTCTCAATCCTAGC TGAAGTCAAATCCCTGCTTC | 196~226 | 58 |
| 1CA32 | TAMRA | AGTTACCAAATGTCGCATTGC TTCATCTGTAAAATGGGCAGG | 107 | 58 |
| 1CA44 | TAMRA | GGCAGCACACCAAATCAAGT TCCTGCAAAACAACAGAGGA | 203 | 58 |
Table 2
mtDNA primers and annealing condition information of E. h. hemionus"
| 基因 Gene | 引物(5'→3') Primer | 扩增条件 Annealing condition |
| CYTB | AACTGCAGTCATCTCCGGTTTACAAGAC CGAAGCTTGATATGAAAAACCATCGTTG | 95 ℃ 4 min; 35 cycles: 95 ℃ 30 s, 50 ℃ 30 s, 72 ℃ 30 s; 72 ℃ 5 min. |
| CYTB* | ATGACAAACATCCGAAAGTC ACTACAGGGACTCTTCACTT | |
| D-LOOP | CTTGTAAACCAGGAAAGGGGGAAAC ATTTAGAGGGCATTCTCACTGGGAT | 95 ℃ 4 min; 35 cycles: 95 ℃ 30 s, 60 ℃ 30 s, 72 ℃ 30 s; 72 ℃ 5 min. |
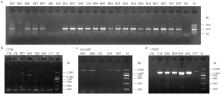
Fig. 1
Agarose gel electrophoresis detection results of PCR amplification using microsatellite and mitochondrial molecular markers in partial samples of Kalamaili population a. Agarose gel electrophoresis result of the amplification at microsatellite locus 10 (300 g·L-1 agarose gel concentration): M. DL 500 molecular weight marker; b, c, d. Agarose gel electrophoresis results of amplification using mitochondrial CYTB, D-LOOP, and CYTB* primers (150 g·L-1 agarose gel concentration): M. DL 2000 molecular weight marker; CK. Negative control used for the batch of fecal genomic DNA extraction"

Table 3
Genetic diversity indicators of E. h. hemionus based on microsatellite molecular markers"
| 位点 Locus | 个体数 N | 等位基因数Na | 有效等位基因数Ne | 香农指数 I | 多态信息含量PIC | 观测杂合度Ho | 期望杂合度He | 近交系数 Fis | 哈迪-温伯格平衡检测 P-value HWE |
| AHT40 | 159 | 22 | 8.032 | 2.391 | 0.864 | 0.711 | 0.875 | 0.159** | 0.000** |
| COR053 | 159 | 20 | 8.689 | 2.387 | 0.874 | 0.736 | 0.885 | 0.109** | 0.016* |
| AHT021 | 159 | 16 | 6.303 | 2.234 | 0.829 | 0.698 | 0.841 | 0.166** | 0.000** |
| HMS2 | 159 | 14 | 5.896 | 1.992 | 0.810 | 0.836 | 0.830 | -0.010 | 0.466 |
| 1CA41 | 159 | 13 | 4.715 | 1.825 | 0.757 | 0.547 | 0.788 | 0.196** | 0.000** |
| 1CA25 | 159 | 10 | 2.079 | 1.087 | 0.483 | 0.321 | 0.519 | 0.323** | 0.000** |
| 1CA43 | 159 | 10 | 1.515 | 0.835 | 0.331 | 0.277 | 0.340 | 0.103* | 0.035* |
| COR082 | 159 | 9 | 3.709 | 1.548 | 0.696 | 0.654 | 0.730 | 0.072 | 0.000** |
| 1CA32 | 159 | 6 | 2.213 | 1.076 | 0.500 | 0.522 | 0.548 | 0.045 | 0.000** |
| 1CA44 | 159 | 3 | 1.283 | 0.393 | 0.198 | 0.025 | 0.221 | 0.826** | 0.000** |
| Mean | 159 | 12.3 | 4.444 | 1.577 | 0.634 | 0.533 | 0.658 | 0.140** | / |
| SE | 0 | 1.88 | 0.858 | 0.222 | 0.080 | 0.075 | 0.080 | / |
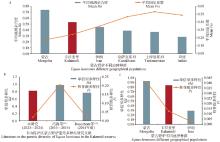
Fig. 2
Comparative analysis of genetic diversity levels in Kalamaili E. h. hemionus population (Data from reference[8-11, 15, 25]) a. Comparison of genetic diversity indices in the E. h. hemionus population based on microsatellite molecular markers; b. Comparative analysis of D-LOOP sequences genetic diversity indices between different periods in the Kalamaili population; c. Comparison of genetic diversity indices in the E. h. hemionus population based on mitochondrial data"

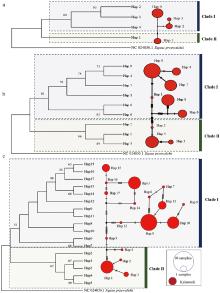
Fig. 3
Genetic structure and phylogenetic relationships of the Kalamaili E. h. hemionus population a. Maximum likelihood (ML) phylogenetic tree and haplotype network of Kalamaili population based on CYTBsequences; b. ML phylogenetic tree and haplotype network of Kalamaili population based on D-LOOPsequences; c. ML phylogenetic tree and haplotype network of Kalamaili population based on combined sequences"

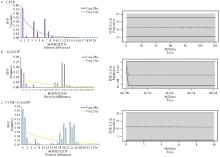
Fig. 7
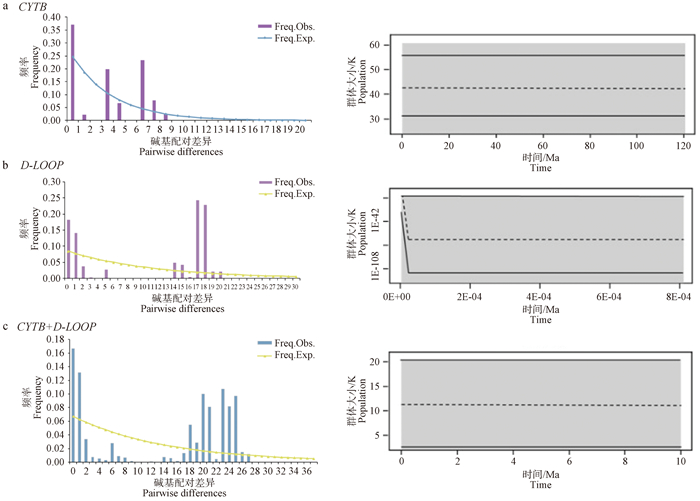
Mismatch distribution and Bayesian skyline plot of the E. h. hemionus population in Kalamaili a. Mismatch distribution and Bayesian skyline plot based on the CYTBgene; b. Mismatch distribution and Bayesian skyline plot based on the D-LOOPgene; c. Mismatch distribution and Bayesian skyline plot based on the combined gene"
| 1 | KACZENSKY P, BAYARBAATAR B, PAYNE J, et al. A conservation strategy for khulan in Mongolia: background and key considerations[R]. NINA Report 1889. Trondheim: Norwegian Institute for Nature Research, 2020. |
| 2 | 中华人民共和国国务院. 国家重点保护野生动物名录[S]. 北京: 中华人民共和国国务院, 2021. |
| The State Council of the people's republic of China. National Key Protected Wildlife List[S]. Beijing: The State Council of the People's Republic of China, 2021. (in Chinese) | |
| 3 | KACZENSKY P, LKHAGVASUREN B, PERELADOVA O, et al. Equus hemionus (amended version of 2015 assessment)[S]. The IUCN Red List of Threatened Species, 2020: eT7951A166520460. |
| 4 | KING S R B, KACZENSKY P. Equus hemionus (Green Status assessment)[S]. The IUCN Red List of Threatened Species 2024: e. T7951A795120251. |
| 5 | XUW,LIUW,MAW,et al.Current status and future challenges for khulan (Equus hemionus) conservation in China[J].Glob Ecol Conserv,2022,37,e02156. |
| 6 | DINGJ J,ZHOUY Y,XUW X,et al.Synergistic effects of climate and land use change on khulan (Equus hemionus hemionus) habitat in China[J].Glob Ecol Conserv,2024,54,123-135. |
| 7 | 张晓晨,邵长亮,葛炎,等.新疆卡拉麦里山有蹄类野生动物自然保护区夏季蒙古野驴适宜生境与种群数量评估[J].应用生态学报,2020,31(9):2993-3004. |
| ZHANGX C,SHAOC L,GEY,et al.Assessment of suitable habitat and population size of khulan (Equus hemionus) in summer in the Kalamaili Mountain Ungulate Nature Reserve, Xinjiang, China[J].Chinese Journal of Applied Ecology,2020,31(9):2993-3004. | |
| 8 |
KACZENSKYP,KOVTENE,HABIBRAKHMANOVR,et al.Genetic characterization of free-ranging Asiatic wild ass in Central Asia as a basis for future conservation strategies[J].Conserv Genet,2018,19(5):1169-1184.
doi: 10.1007/s10592-018-1086-3 |
| 9 |
DEVENDRAK,ASHWINA,SAMEERAF,et al.Mitochondrial DNA analyses revealed low genetic diversity in the endangered Indian wild ass Equus hemionus khur[J].Mitochondrial DNA, Part A,2017,28(5):681-686.
doi: 10.3109/24701394.2016.1174221 |
| 10 | DEVENDRAK,ASHWINA,SAMEERAF,et al.Low genetic diversity of the endangered Indian wild ass Equus hemionus khur, as revealed by microsatellite analyses[J].J Genet,2017,96(2):e31-34. |
| 11 | ROSENBOMS,COSTAV,CHENS,et al.Reassessing the evolutionary history of ass-like equids: Insights from patterns of genetic variation in contemporary extant populations[J].Mol Phylogenet Evol,2015,8588-8596. |
| 12 |
BENNETTA E,CHAMPLOTS,PETERSJ,et al.Taming the late quaternary phylogeography of the Eurasiatic wild ass through ancient and modern DNA[J].PLoS One,2017,12(4):e0174216.
doi: 10.1371/journal.pone.0174216 |
| 13 | OAKENFULLE A,LIMH N,RYDERO A,et al.A survey of equid mitochondrial DNA: Implications for the evolution, genetic diversity and conservation of Equus[J].Conserv Genet,2000,341-355. |
| 14 |
VILSTRUPJ T,ANDAINEA O,STILLERM,et al.Mitochondrial phylogenomics of modern and ancient equids[J].PLoS One,2013,8(2):e55950.
doi: 10.1371/journal.pone.0055950 |
| 15 | 冯锦,初雯雯,端肖楠,等.新疆卡拉麦里山有蹄类自然保护区蒙古野驴mtDNA D-LOOP区的遗传多样性及系统发育研究[J].野生动物学报,2018,39(4):737-744. |
| FENGJ,CHUW W,DUANX N,et al.Genetic diversity and the phylogenetic status of the mtDNA D-LOOP of Khulan (Equus hemionus) at the Mountain Kalamaili Ungulate Nature Reserve, Xinjiang[J].Wildlife Science Bulletin,2018,39(4):737-744. | |
| 16 | 魏辅文,马天笑,胡义波.中国濒危兽类保护遗传学研究进展与展望[J].兽类学报,2021,41(5):571-580. |
| WEIF W,MAT X,HUY B.Research advances and perspectives of conservation genetics of threatened mammals in China[J].Acta Theriologica Sinica,2021,41(5):571-580. | |
| 17 |
CUIL Y,LIUB Y,LIH M,et al.A simple and effective method to enrich endogenous DNA from mammalian faeces[J].Mol Ecol Resour,2024,24(4):e13939.
doi: 10.1111/1755-0998.13939 |
| 18 | 褚佳宁,徐海涛,何志健,等.我国圈养和野生东北虎种群线粒体基因组遗传多样性的比较研究[J].野生动物学报,2024,45(2):231-241. |
| CHUJ N,XUH T,HEZ J,et al.A comparative Study on mitochondrial genetic diversity between captive and wild Amur Tigers in China[J].Wildlife Science Bulletin,2024,45(2):231-241. | |
| 19 | 周颖娜,李金霖,马跃,等.基于微卫星标记的雪豹种群结构及遗传多样性[J].野生动物学报,2024,45(4):699-708. |
| ZHOUY N,LIJ L,MAY,et al.Population structure and genetic diversity of snow leopard based on microsatellite markers[J].Wildlife Science Bulletin,2024,45(4):699-708. | |
| 20 | YINQ Q,RENZ,WENX Y,et al.Assessment of population genetic diversity and genetic structure of the North Chinese leopard (Panthera pardus japonensis) in fragmented habitats of the Loess Plateau, China[J].Glob Ecol Conserv,2023,42,e02416. |
| 21 |
KATEA,JAPNINGJ R R,GIARATN A N A,et al.Emerging patterns of genetic diversity in the critically endangered Malayan tiger (Panthera tigris jacksoni)[J].Biodivers Conserv,2024,33(4):1325-1349.
doi: 10.1007/s10531-024-02799-9 |
| 22 | 李威. 海南长臂猿Nomascus hainanus种群扩大后的遗传变化分析[D]. 贵州: 贵州师范大学, 2024. |
| LI W. Genetic changes of hainan gibbon Nomascus hainanus after population expansion[D]. Guizhou: Guizhou Normal University, 2024. (in Chinese) | |
| 23 | 王思维. 黔金丝猴(Rhinopithecus brelichi)种群遗传学研究[D]. 贵州: 贵州师范大学, 2022. |
| WANG S W. Population genetics study of Rhinopithecus brelichi[D]. Guizhou: Guizhou Normal University, 2022. (in Chinese) | |
| 24 | ZHANGL,SUNG,AZHANHANE,et al.Genetic diversity assessment for reintroduced Przewalski's horse (Equus ferus) based on newly developed SSR markers[J].Conserv Genet Resour,2024,16(1):89-101. |
| 25 |
KACZENSKYP,KUEHNR,LHAGVASURENB,et al.Connectivity of the Asiatic wild ass population in the Mongolian Gobi[J].Biol Conserv,2011,144(2):920-929.
doi: 10.1016/j.biocon.2010.12.013 |
| 26 | 新疆维吾尔自治区林业和草原局. 关于新疆卡拉麦里山有蹄类野生动物自然保护区面积范围及功能分区的函[S]. 乌鲁木齐: 新疆维吾尔自治区林业和草原局, 2024. |
| Xinjiang Uygur Autonomous Region Forestry and Grassland Bureau. Letter on the area scope and functional zoning of the Xinjiang Kalamaili Ungulate Nature Reserve [S]. Urumqi: Xinjiang Uygur Autonomous Region Forestry and Grassland Bureau, 2024. (in Chinese) | |
| 27 | 新疆维吾尔自治区林业和草原局. 新疆卡拉麦里山自然保护区尽显生态之美[R]. 乌鲁木齐: 新疆维吾尔自治区林业和草原局, 2024. |
| Xinjiang Uygur Autonomous Region Forestry and Grassland Bureau. The ecological beauty of Xinjiang Kalamaili Nature Reserve[R]. Urumqi: Xinjiang Uygur Autonomous Region Forestry and Grassland Bureau, 2024. (in Chinese) | |
| 28 |
CHOWDHARYB P,RAUDSEPPT,KATAS R,et al.The first-generation whole-genome radiation hybrid map in the horse identifies conserved segments in human and mouse genomes[J].Genome Res,2003,13(4):742-751.
doi: 10.1101/gr.917503 |
| 29 | 杨方园, 巫鹏翔. 微卫星标记的开发和数据分析流程[R]. |
| Bio-101 e1010608, 2021. YANG F Y, WU P Y. Protocol for Development, Genotyping and Data Analysis of Microsatellite Maker[R]. Bio-101 e1010608, 2021. (in Chinese) | |
| 30 |
STEINERC C,RYDERA O.Molecular phylogeny and evolution of the Perissodactyla[J].Zool J Linn Soc,2011,163(4):1289-1303.
doi: 10.1111/j.1096-3642.2011.00752.x |
| 31 |
ITOH Y,LANGENHORSTT,OGDENR,et al.Population genetic diversity and hybrid detection in captive zebras[J].Sci Rep,2015,5(1):e13171.
doi: 10.1038/srep13171 |
| 32 |
KOTZEA,SMITHR M,MOODLEYY,et al.Lessons for conservation management: Monitoring temporal changes in genetic diversity of cape mountain zebra (Equus zebra zebra)[J].PloS one,2019,14(7):e0220331.
doi: 10.1371/journal.pone.0220331 |
| 33 |
SHIL P,YANGX F,CHAM H,et al.Genetic diversity and structure of mongolian gazelle (Procapra gutturosa) populations in fragmented habitats[J].BMC genomics,2023,24(1):507.
doi: 10.1186/s12864-023-09574-0 |
| 34 | 李峥. 大兴安岭南麓东北马鹿遗传结构及景观与遗传分化格局的关系[D]. 黑龙江: 东北林业大学, 2023. |
| LI Z. Genetic structure of red deer and the relationship between landscape and genetic differentiation pattern in the southern of Greater Khingan Mountains, China[D]. Heilongjiang: Northeast Forestry University, 2023. (in Chinese) | |
| 35 |
DUY R,ZOUX Y,XUY T,et al.Microsatellite Loci Analysis Reveals Post-bottleneck recovery of genetic diversity in the tibetan antelope[J].Sci Rep,2016,6,e35501.
doi: 10.1038/srep35501 |
| 36 | FANUELK,SONIAR,LEILIK,et al.Genetic diversity of the Ethiopian Grevy′s zebra (Equus grevyi) populations that includes a unique population of the Alledeghi Plain[J].Mitochondrial DNA A DNA Mapp Seq Anal,2016,27(1):397-400. |
| 37 |
MOODLEYY S,HARLEYH E.Population structuring in mountain zebras (Equus zebra): The molecularconsequences of divergent demographic histories[J].Conserva Genet,2006,6(6):953-968.
doi: 10.1007/s10592-005-9083-8 |
| 38 |
ELINED L,PETERA,HANSR S.High variation and very low differentiation in wide ranging plains zebra (Equus quagga): Insights from mtDNA and microsatellites[J].Mol Ecol,2008,17(12):2812-2824.
doi: 10.1111/j.1365-294X.2008.03781.x |
| 39 |
AHMADK,KUMARVP,JOSHIB D,et al.Genetic diversity of the Tibetan antelope (Pantholops hodgsonii) population of Ladakh, India, its relationship with other populations and conservation implications[J].BMC Res Notes,2016,9(1):e477.
doi: 10.1186/s13104-016-2271-4 |
| 40 | 董潭成,初红军,陈勇,等.新疆卡拉麦里山有蹄类自然保护区鹅喉羚遗传多样性及系统发育地位[J].兽类学报,2016,36(1):77-86. |
| DONGT C,CHUH J,CHENY,et al.Genetic diversity and phylogenetic status of Gazella subgutturosa at the Mountain Kalamaili Ungulate Nature Reserve, Xinjiang[J].Acta Theriologica Sinica,2016,36(1):77-86. | |
| 41 |
米热姑丽·麦麦提,周世玉,刘鹏,等.基于线粒体基因的新疆托氏兔遗传多样性和种群遗传结构评价[J].畜牧兽医学报,2022,53(1):112-121.
doi: 10.11843/j.issn.0366-6964.2022.01.011 |
|
MAMAT M,ZHOUS Y,LIUP,et al.Evaluation of genetic diversity and population genetic structure of Tolai Hare in Xinjiang based on mitochondrial DNA[J].Acta Veterinaria et Zootechnica Sinica,2022,53(1):112-121.
doi: 10.11843/j.issn.0366-6964.2022.01.011 |
|
| 42 | SHAWE R,FARQUHARSONA K,BRUFORDW M,et al.Global meta-analysis shows action is needed to halt genetic diversity loss[J].Nature,2025(prepublish):1-7. |
| 43 |
JHALAV Y,MUNGIA N,GOPALR,et al.Tiger recovery amid people and poverty[J].Science,2025,387(6733):505-510.
doi: 10.1126/science.adk4827 |
| 44 |
LUOY,CHENY,LIUF,JIANGC,et al.Mitochondrial genome sequence of the Tibetan wild ass (Equus kiang)[J].Mitochondrial DNA,2011,22(1-2):6-8.
doi: 10.3109/19401736.2011.588221 |
| 45 |
MUSTAFAO,KANATG,ERENY,et al.The first complete genome of the extinct European wild ass (Equus hemionus hydruntinus)[J].Mol Ecol,2024,33(14):e17440.
doi: 10.1111/mec.17440 |
| 46 | 初红军,蒋志刚,葛炎,等.卡拉麦里山有蹄类自然保护区蒙古野驴和鹅喉羚种群密度和数量[J].生物多样性,2009,17(4):414-422. |
| CHUH J,JIANGZ G,GEY,et al.Population density and abundance of khulan (Equus hemionus) and goitered gazelle (Gazella subgutturosa) in the Kalamaili Mountain Ungulate Nature Reserve[J].Biodiversity Science,2009,17(4):414-422. | |
| 47 | 初雯雯. 新疆卡山自然保护区蒙古野驴和鹅喉羚资源现状和社区牧民保护意识研究[D]. 北京: 北京林业大学, 2019. |
| CHU W W. Resource status of khulan (Equus hemionus) and goitered gazelle (Gazella subgutturosa) and conservation awareness of local herders in the Kalamaili Mountain Nature Reserve, Xinjiang [D]. Beijing: Beijing Forestry University, 2019. (in Chinese) | |
| 48 | 陈艳秋,初雯雯,李基才,等.新疆卡拉麦里山有蹄类自然保护区夏季鹅喉羚和蒙古野驴的生境适宜性和重叠性分析[J].四川动物,2024,43(3):264-273. |
| CHENY Q,CHUW W,LIJ C,et al.Habitat suitability and overlap analysis of goitered gazelle (Gazella subgutturosa) and khulan (Equus hemionus) in summer in the Kalamaili Mountain Ungulate Nature Reserve, Xinjiang[J].Sichuan Journal of Zoology,2024,43(3):264-273. |
| [1] | LIU Sha, YANG Caichun, ZHANG Xiaoyu, CHEN Qiong, LIU Xiong, CHEN Hongbo, ZHOU Huanhuan, SHI Liangyu. Population Genetic Structure and Genome-wide Runs of Homozygosity Analysis in Meihuaxing Pigs Based on 80K SNP Chip [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(8): 3749-3760. |
| [2] | REN Qianzi, ZHANG Baizhong, WANG Zhenqing, WANG Xianglin, GONG Ying, HU Renke, PU Yabin, SU Peng, LI Yefang, MA Yuehui, LI Haobang, JIANG Lin. Genetic Evolutionary Analysis of Wuxue Goat Based on Whole Genome Resequencing [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(8): 3787-3801. |
| [3] | ZHANG Jialiang, HUANG Chang, YANG Yonglin, YANG Hua, BAI Wenlin, MA Yuehui, ZHAO Qianjun. Genetic Structure and Wool Trait Selection Signatures Analysis of Chinese Sheep Populations Based on 50K Liquid SNP Chip [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(7): 3164-3176. |
| [4] | MIAO Junjie, ZHANG Riquan, WU Houyi, YOU Xinming, HUANG Yiwen, HUANG Xiaoying, GUO Zhenyang, LIU Jianlin, XIAO Weihua, GUO Tianhua, CHEN Hao, KANG Dongliu. Genome-Wide SNP Analysis Revealed the Characteristics of Germplasm Resources and Genetic Diversity of Jinggang Black-Palm Geese [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(7): 3199-3209. |
| [5] | WANG Qinqian, GAO Zhendong, LU Ying, MA Ruoshan, DENG Weidong, HE Xiaoming. Research Progress of Whole Genome Resequencing in Chinese Indigenous Cattle [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(5): 2026-2037. |
| [6] | YAO Tingting, LI Hao, YAN Huixuan, CAO Yifan, Cirengluobu , Suolangquji , Nimacangjue , ZHAO Li, Danzengluosang , Silangwangmu , Basangzhuzha , CHEN Ningbo. Genetic Diversity of Mitochondrial Genome and Maternal Origin of 10 Cattle Populations in Tibet Autonomous Region [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(5): 2194-2202. |
| [7] | WANG Haoyu, MA Keyan, LI Taotao, LI Dengpan, ZHAO Qing, MA Youji. Population Genetic Diversity and Population Structure Analysis of Small-boned Goat Based on Specific-Locus Amplified Fragment Sequencing [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(3): 1170-1179. |
| [8] | HU Xin, YOU Wei, JIANG Fugui, CHENG Haijian, SUN Zhigang, SONG Enliang. Analysis of Genetic Diversity and Population Structure of Simmental Cattle Based on Whole Genome Resequencing [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(3): 1189-1202. |
| [9] | WAN Weican, HE Xu, LIU Yang, MA Yuyong, JIANG Yuzhang, DAI Qiuzhong, YAN Haifeng, JIANG Guitao, LI Chuang. Evaluation of Daozhou Gray Goose Conservation Based on Whole Genome Resequencing Analysis [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(2): 633-642. |
| [10] | LIANG Entang, LI Huaxuan, CHEN Shuaicheng, LI Guo, SUN Gege, ZAN Linsen. Effect of Genistein on Semen Cryopreservation of Bull [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(2): 700-710. |
| [11] | FENG Da, WEI Chen, HU Siyi, DU Chunmei, MA Jian, WU Jiang, ZHOU Guangxian, GAN Shangquan. Genetic Diversity and Population Structure Analysis of Leizhou Goat Based on Whole Genome Resequencing Analysis [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(10): 4947-4962. |
| [12] | Shuo YANG, Min HUO, Zixuan SU, Yuxiang SHI. Research Progress on the Impact of Mitochondrial Quality Control on Oxidative Stress in Livestock and Poultry [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(9): 3769-3776. |
| [13] | Siyu LIU, Man ZHANG, Yan ZHANG, Zhitong WEI, Xinglei QI, Tengyun GAO, Xian LIU, Dong LIANG, Tong FU. Evaluation of the Conservation Effect in Nanyang Cattle Based on Resequencing Data [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(9): 3876-3886. |
| [14] | Hongyan HUANG, Liyun ZHANG, Zhirong HUANG, Zhongping WU, Xumeng ZHANG, Hongjia OUYANG, Junpeng CHEN, Zhenping LIN, Yunbo TIAN, Xiujin LI, Yunmao HUANG. The Study on Population Genetic Diversity and Genome-wide Association Study of Body Weight and Size Traits for Lion-head Geese [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(9): 3914-3924. |
| [15] | Yi WANG, Jianfei GONG, Nuo HENG, Yingfan HU, Rui WANG, Huan WANG, Ni ZHU, Wei HE, Zhihui HU, Haisheng HAO, Huabin ZHU, Shanjiang ZHAO. Melatonin Alleviates Palmitic Acid-induced Damage in Bovine Endometrial Epithelial Cells by Improving Mitochondrial Dynamics [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(9): 3978-3987. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||