





Acta Veterinaria et Zootechnica Sinica ›› 2024, Vol. 55 ›› Issue (11): 4950-4967.doi: 10.11843/j.issn.0366-6964.2024.11.015
• Animal Genetics and Breeding • Previous Articles Next Articles
Ting JIANG1( ), Wendong LI1, Xingqi LI1, Yu HUANG1, Qigui WANG2, Haiwei WANG2, Chaowu YANG3, Lingbin LIU1,*(
), Wendong LI1, Xingqi LI1, Yu HUANG1, Qigui WANG2, Haiwei WANG2, Chaowu YANG3, Lingbin LIU1,*( )
)
Received:2024-05-13
Online:2024-11-23
Published:2024-11-30
Contact:
Lingbin LIU
E-mail:jiangt019@163.com;liulb515@163.com
CLC Number:
Ting JIANG, Wendong LI, Xingqi LI, Yu HUANG, Qigui WANG, Haiwei WANG, Chaowu YANG, Lingbin LIU. Screening Candidate Genes for Ovarian Development and Constructing Regulatory Network in Nesting Chickens by Transcriptome and Proteome[J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(11): 4950-4967.
Table 1
Primers sequence for qRT-PCR"
| 基因Gene | 上游引物(5′→3′) Forward primer | 下游引物(3′→5′) Reverse primer | 产物长度/bp Product length |
| FN1 | CACCTACCCCCAGTTCACAC | TGGATGTTGTGTGTCTGCCT | 109 |
| COL1A1 | GCCAAGACGAAGACATCCCA | GGCAGTTCTTGGTCTCGTCA | 156 |
| SGK2 | GCCAAACGCAAGAGTGATGG | TCTCCGAGGTCTGGAAGGAG | 169 |
| CASTOR1 | CTGTGTCCTCACACTGGACC | AACGTGATGGAGCTGGGTTC | 129 |
| ACAT1 | GGTTCTGATGACCACGGAGG | GAATCTTGGGAACAGCGTGC | 131 |
| MMP10 | CTCTATGGACCCCCAAGGGA | TGGGTGCCATTTCTGTTGGT | 121 |
| β-actin | CTGTGCCCATCTATGAAGGCTA | ATTTCTCTCTCGGCTGTGGTG | 132 |

Fig. 1
Observation of ovarian tissue anatomy and HE staining of Chengkou mountain chicken A. Status of Chengkou mountain chicken at normal laying period; B. Status of the Chengkou mountain chicken at nesting stage; C. Developing ovary that continuously produce eggs; D. Atrophic ovary on the 10 days of nesting; E. Atrophic ovary on the 20 days of nesting; F. Atrophic ovary on the 30 days of nesting; G. HE staining sections of ovarian tissues in NO group; H. HE staining sections of ovarian tissues in AO group"

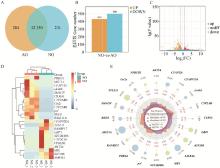
Fig. 2
Differential expression analysis of mRNAs A. mRNAs expression statistics; B. Statistics of up-regulated and down-regulated differentially expressed mRNAs; C. Volcano plot of differentially expressed mRNAs; D. Clustering heatmap of differentially expressed mRNAs; E. Radar plot of the top 20 differentially expressed genes with the largest |log2FC| value"

Table 2
Top 20 upregulated DEGs in broodiness of chickens atrophy ovaries"
| 基因Gene | 描述Description | log2(FC) | P-value |
| TTLL11 | Tubulin polyglutamylase TTLL11 isoform X4 | 10.500 841 88 | 1.458 15×10-5 |
| VPREB3 | Pre-b lymphocyte 3 | 6.516 213 893 | 0.000 819 366 |
| AICDA | Activation induced cytidine deaminase | 5.560 132 636 | 0.001 782 563 |
| Aldh3b1 | Aldehyde dehydrogenase family 3 member b1 | 5.347 768 100 | 0.000 477 674 |
| CXCL13 | C-x-c motif chemokine ligand 13 | 4.643 001 006 | 0.010 083 317 |
| CHIR-B2 | Gallus gallus immunoglobulin-like receptor chir-b2 | 4.070 460 591 | 0.000 434 750 |
| MR1 | F10 alpha chain-like isoform x1 | 3.665 580 961 | 0.016 924 252 |
| PAX5 | Paired box 5 | 3.554 588 852 | 0.034 263 113 |
| CXCR5 | C-x-c motif chemokine receptor 5 | 3.372 478 676 | 0.000 260 851 |
| SLC6A18 | Solute carrier family 6 member 18 | 3.145 381 031 | 0.000 118 215 |
| CD79B | Cd79b molecule | 3.030 284 866 | 5.085 67×10-5 |
| RGS13 | Regulator of g-protein signaling 13 | 3.000 836 587 | 0.014 083 046 |
| BPTF | Bromodomain phd finger transcription factor, partial | 2.964 106 33 | 0.000 817 016 |
| pol | Uncharacterized protein loc106895380 | 2.701 115 657 | 0.040 790 927 |
| sebox | Paired box protein 6 homolog | 2.626 303 296 | 0.000 660 700 |
| RGS20 | Regulator of g-protein signaling 20 | 2.588 546 838 | 0.000 265 790 |
| Tnr | Fibrinogen-like protein 1-like protein | 2.567 345 957 | 4.961 03×10-5 |
| SLC22A18 | Solute carrier family 22 member 18 | 2.460 845 463 | 0.000 527 036 |
| GSX2 | Gs homeobox 2-like | 2.448 250 869 | 0.012 140 053 |
| POU2AF1 | Pou class 2 associating factor 1 | 2.421 874 929 | 0.002 290 982 |
Table 3
Top 20 downregulated DEGs in broodiness of chicken atrophy ovaries"
| 基因Gene | 描述Description | log2(FC) | P-value |
| CYP2AB1 | Cytochrome P450, family 2, subfamily AB, polypeptide 1 | -5.805 985 902 | 1.147 4×10-151 |
| ACTC1 | Actin alpha cardiac muscle 1 | -4.874 610 515 | 1.073 13×10-7 |
| ALB | Albumin | -4.060 938 705 | 4.069 68×10-6 |
| UMOD | Uromodulin | -4.015 674 232 | 6.976 03×10-8 |
| ZP3 | Zona pellucida sperm-binding protein 3 | -3.809 355 324 | 5.409 94×10-10 |
| COL8A1 | Collagen type Ⅷ alpha 1 chain | -3.743 850 285 | 3.122 75×10-9 |
| ACTA1 | Actin alpha 1, skeletal muscle | -3.444 999 220 | 6.556 52×10-10 |
| MMP13 | Matrix metallopeptidase 13 | -3.430 684 692 | 7.028 18×10-5 |
| TAGLN | Transgelin | -3.213 487 543 | 2.005 09×10-11 |
| MYL4 | Myosin, light chain 4, alkali; atrial, embryonic | -3.204 401 269 | 2.002 51×10-13 |
| FAM189A2 | Family with sequence similarity 189 member A2 | -3.123 454 143 | 5.063 02×10-7 |
| INHA | Inhibin alpha subunit | -3.115 492 881 | 7.802 8×10-5 |
| MMP10 | Matrix metallopeptidase 10 | -3.045 843 231 | 1.819 7×10-27 |
| ACTG2 | Actin, gamma 2, smooth muscle, enteric | -2.980 748 271 | 1.992 53×10-9 |
| CNN1 | Calponin 1 | -2.957 732 234 | 3.116 93×10-23 |
| SFRP4 | Secreted frizzled related protein 4 | -2.826 452 510 | 1.254 79×10-19 |
| COL4A1 | Collagen type Ⅳ alpha 1 chain | -2.821 702 234 | 1.732 3×10-25 |
| VCAN | Versican | -2.761 868 293 | 4.999 09×10-36 |
| COL4A2 | Collagen type Ⅳ alpha 2 chain | -2.702 337 677 | 1.318 83×10-21 |
| ACTA2 | Actin, alpha 2, smooth muscle, aorta | -2.687 926 646 | 1.802 76×10-6 |
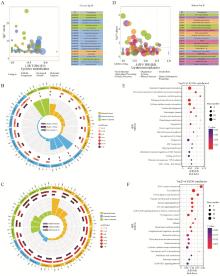
Fig. 3
Enrichment analysis of DEGs A. Bubble plot of GO enrichment analysis of DEGs; B. GO enrichment analysis circle diagram of up-regulated DEGs; C. GO enrichment analysis circle diagram of down-regulated DEGs; D. Bubble plots of KEGG pathway enrichment analysis of DEGs; E. Bubble plots of KEGG enrichment analysis of up-regulated DEGs; F. Bubble plot of KEGG enrichment analysis of down-regulated DEGs"


Fig. 4
Analysis of regulators of PI3K-Akt signaling pathway A. Experimental protocol for identification of regulators involved in PI3K-Akt signaling pathway; B. GSEA analysis of cell growth and death-related pathways; C. GSEA enrichment map of PI3K-Akt signaling pathway; D. Heatmap of expression levels of 23 DEGs in PI3K-Akt signaling pathway; E. PPI co-expression network of 23 DEGs; F. GO enrichment analysis of PI3K-Akt signaling pathway regulators; G. KEGG enrichment analysis of PI3K-Akt signaling pathway regulators"


Fig. 5
Differential expression analysis of proteins A. Protein expression statistics; B. Statistics of up-regulated and down-regulated differentially expressed proteins; C. Volcano plot of differentially expressed proteins; D. Top 20 differentially expressed proteins with the largest |log2FC| value; E. Clustering heatmap of differentially expressed proteins"

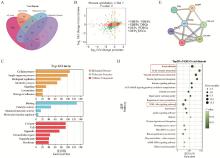
Fig. 7
Combined analysis of transcriptomics and proteomics A. Bubble plot of GO enrichment analysis of DEPs; B. Four-quadrant diagram; C. GO enrichment analysis of DEPs_DEGs: Different colors represent different types of GO terms; D. KEGG pathway analysis of DEPs_DEGs: The top 20 enrichment pathways; E. PPI network diagram of candidate signaling pathways"

Table 4
KEGG analysis of the DEPs and DEGs"
| KEGG通路KEGG pathway | 差异表达基因、差异表达蛋白DEGs, DEPs | 基因数Gene number |
| Focal adhesion | MYLK、COMP、FN1、ITGA8、THBS1、COL4A2、MYL9、COL4A1、TNC、COL1A1、ACTN1、FLNA | 12 |
| ECM-receptor interaction | COMP、FN1、ITGA8、THBS1、SDC1、COL4A2、COL4A1、TNC、COL1A1 | 9 |
| PI3K-Akt signaling pathway | COMP、SGK2、FN1、ITGA8、THBS1、COL4A2、EIF4E1B、COL4A1、TNC、COL1A1 | 10 |
| 1 |
李俊, 李建伟, 杨蓉, 等. 家禽就巢性影响因素的研究进展[J]. 中国畜禽种业, 2023, 19 (8): 91- 95.
doi: 10.3969/j.issn.1673-4556.2023.08.018 |
|
LI J , LI J W , YANG R , et al. Research progress on factors affecting broodiness in poultry[J]. The Chinese Livestock and Poultry Breeding, 2023, 19 (8): 91- 95.
doi: 10.3969/j.issn.1673-4556.2023.08.018 |
|
| 2 |
ROMANOV M N , TALBOT R T , WILSON P W , et al. Genetic control of incubation behavior in the domestic hen[J]. Poult Sci, 2002, 81 (7): 928- 931.
doi: 10.1093/ps/81.7.928 |
| 3 | 刘安怀, 徐茂森, 张旭, 等. 产蛋期和就巢期皖西白鹅卵巢组织结构、血清生化指标和激素水平比较研究[J]. 安徽科技学院学报, 2023, 37 (2): 1- 5. |
| LIU A H , XU M S , ZHANG X , et al. Effects of brooding behavior on ovarian tissue structure, blood biochemical indexes and hormone levels of Wanxi white geese[J]. Journal of Anhui Science and Technology University, 2023, 37 (2): 1- 5. | |
| 4 | 王思博, 杨季, 马渭青, 等. 鹅就巢期卵巢特征及其内分泌机制的研究进展[J]. 中国畜牧杂志, 2020, 56 (3): 33- 37. |
| WANG S B , YANG J , MA W Q , et al. Characteristics of ovary and its endocrine mechanism in geese during nesting[J]. Chinese Journal of Animal Science, 2020, 56 (3): 33- 37. | |
| 5 |
ZHAO J B , PAN H B , LIU Y , et al. Interacting networks of the hypothalamic-pituitary-ovarian axis regulate layer hens performance[J]. Genes (Basel), 2023, 14 (1): 141.
doi: 10.3390/genes14010141 |
| 6 |
YU J , LOU Y P , HE K , et al. Goose broodiness is involved in granulosa cell autophagy and homeostatic imbalance of follicular hormones[J]. Poult Sci, 2016, 95 (5): 1156- 1164.
doi: 10.3382/ps/pew006 |
| 7 |
LIU L B , XIAO Q H , GILBERT E R , et al. Whole-transcriptome analysis of atrophic ovaries in broody chickens reveals regulatory pathways associated with proliferation and apoptosis[J]. Sci Rep, 2018, 8 (1): 7231.
doi: 10.1038/s41598-018-25103-6 |
| 8 |
JOHNSON A L , BRIDGHAM J T , WITTY J P , et al. Expression of bcl-2 and nr-13 in hen ovarian follicles during development[J]. Biol Reprod, 1997, 57 (5): 1096- 1103.
doi: 10.1095/biolreprod57.5.1096 |
| 9 |
XU G Q , DONG Y Y Y , WANG Z , et al. Melatonin Attenuates Oxidative Stress-induced apoptosis of bovine ovarian granulosa cells by promoting mitophagy via SIRT1/FoxO1 signaling pathway[J]. Int J Mol Sci, 2023, 24 (16): 12854.
doi: 10.3390/ijms241612854 |
| 10 |
GARRETT W M , GUTHRIE H D . Steroidogenic enzyme expression during preovulatory follicle maturation in pigs[J]. Biol Reprod, 1997, 56 (6): 1424- 1431.
doi: 10.1095/biolreprod56.6.1424 |
| 11 |
SHEN X , BAI X , LUO C L , et al. Quantitative proteomic analysis of chicken serum reveals key proteins affecting follicle development during reproductive phase transitions[J]. Poult Sci, 2021, 100 (1): 325- 333.
doi: 10.1016/j.psj.2020.09.058 |
| 12 |
JIANG R S , XU G Y , ZHANG X Q , et al. Association of polymorphisms for prolactin and prolactin receptor genes with broody traits in chickens[J]. Poult Sci, 2005, 84 (6): 839- 845.
doi: 10.1093/ps/84.6.839 |
| 13 |
PONSUKSILI S , TRAKOOLJUL N , HADLICH F , et al. Genetic regulation of liver metabolites and transcripts linking to biochemical-clinical parameters[J]. Front Genet, 2019, 10, 348.
doi: 10.3389/fgene.2019.00348 |
| 14 |
JIANG S S , ZHANG G H , MIAO J , et al. Transcriptome and metabolome analyses provide insight into the glucose-induced adipogenesis in porcine adipocytes[J]. Curr Issues Mol Biol, 2024, 46 (3): 2027- 2042.
doi: 10.3390/cimb46030131 |
| 15 | 王亚鑫, 王璟, 田学凯, 等. 多组学技术在畜禽重要经济性状研究中的应用[J]. 畜牧兽医学报, 2024, 55 (5): 1842- 1853. |
| WANG Y X , WANG J , TIAN X K , et al. Application of multi-omics technology in the study of important economic traits of livestock and poultry[J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55 (5): 1842- 1853. | |
| 16 | HUANG X , ZHANG H Y , CAO H Y , et al. Transcriptomics and metabolomics analysis of the ovaries of high and low egg production chickens[J]. Animals (Basel), 2022, 12 (16): 2010. |
| 17 |
WANG L Y , ZHANG Y W , ZHANG B , et al. Candidate gene screening for lipid deposition using combined transcriptomic and proteomic data from Nanyang black pigs[J]. BMC Genomics, 2021, 22 (1): 441.
doi: 10.1186/s12864-021-07764-2 |
| 18 |
TAO Z , SONG W , ZHU C , et al. Comparative transcriptomic analysis of high and low egg-producing duck ovaries[J]. Poult Sci, 2017, 96 (12): 4378- 4388.
doi: 10.3382/ps/pex229 |
| 19 |
SHEN X , BAI X , XU J , et al. Transcriptome sequencing reveals genetic mechanisms underlying the transition between the laying and brooding phases and gene expression changes associated with divergent reproductive phenotypes in chickens[J]. Mol Biol Rep, 2016, 43 (9): 977- 989.
doi: 10.1007/s11033-016-4033-8 |
| 20 |
KOVÁCS J , FORGÓ V , PÉCZELY P . The fine structure of the follicular cells in growing and atretic ovarian follicles of the domestic goose[J]. Cell Tissue Res, 1992, 267 (3): 561- 569.
doi: 10.1007/BF00319379 |
| 21 |
YU J , LOU Y P , ZHAO A Y . Transcriptome analysis of follicles reveals the importance of autophagy and hormones in regulating broodiness of Zhedong white goose[J]. Sci Rep, 2016, 6 (1): 36877.
doi: 10.1038/srep36877 |
| 22 |
POWERS R K , GOODSPEED A , PIELKE-LOMBARDO H , et al. GSEA-InContext: identifying novel and common patterns in expression experiments[J]. Bioinformatics, 2018, 34 (13): i555- i564.
doi: 10.1093/bioinformatics/bty271 |
| 23 |
HE H R , LI D M , TIAN Y T , et al. miRNA sequencing analysis of healthy and atretic follicles of chickens revealed that miR-30a-5p inhibits granulosa cell death via targeting Beclin1[J]. J Anim Sci Biotechnol, 2022, 13 (1): 55.
doi: 10.1186/s40104-022-00697-0 |
| 24 |
TILLY J L , KOWALSKI K I , JOHNSON A L , et al. Involvement of apoptosis in ovarian follicular atresia and postovulatory regression[J]. Endocrinology, 1991, 129 (5): 2799- 2801.
doi: 10.1210/endo-129-5-2799 |
| 25 |
CHOI J , JO M , LEE E , et al. AKT is involved in granulosa cell autophagy regulation via mTOR signaling during rat follicular development and atresia[J]. Reproduction, 2014, 147 (1): 73- 80.
doi: 10.1530/REP-13-0386 |
| 26 |
KAIPIA A , HSUEH A J W . Regulation of ovarian follicle atresia[J]. Annu Rev Physiol, 1997, 59, 349- 363.
doi: 10.1146/annurev.physiol.59.1.349 |
| 27 | 茹盟, 曾文惠, 彭剑玲, 等. 蛋鸡卵泡发育及其表观遗传调控机制研究进展[J]. 畜牧兽医学报, 2023, 54 (9): 3613- 3622. |
| RU M , ZENG W H , PENG J L , et al. Research progress on follicles development of hens and its epigenetic regulatory mechanism[J]. Acta Veterinaria et Zootechnica Sinica, 2023, 54 (9): 3613- 3622. | |
| 28 | 郑言, 曹中赞, 马贲, 等. 氧化应激对家禽卵泡颗粒细胞凋亡和自噬的影响及机制研究进展[J]. 中国畜牧杂志, 2023, 59 (9): 110- 115. |
| ZHENG Y , CAO Z Z , MA B , et al. Research progress on the effect of oxidative stress on apoptosis and autophagy of poultry follicular granulosa cells and its mechanism[J]. Chinese Journal of Animal Science, 2023, 59 (9): 110- 115. | |
| 29 |
ZHANG Y , ZHANG N J , ZOU Y M , et al. Deacetylation of Septin4 by SIRT2 (silent mating type information regulation 2 Homolog-2) mitigates damaging of hypertensive nephropathy[J]. Circ Res, 2023, 132 (5): 601- 624.
doi: 10.1161/CIRCRESAHA.122.321591 |
| 30 |
YUE J C , LÓPEZ J M . Understanding MAPK signaling pathways in apoptosis[J]. Int J Mol Sci, 2020, 21 (7): 2346.
doi: 10.3390/ijms21072346 |
| 31 |
ZHU G Y , MAO Y , ZHOU W D , et al. Dynamic changes in the follicular transcriptome and promoter DNA methylation pattern of steroidogenic genes in chicken follicles throughout the ovulation cycle[J]. PLoS One, 2015, 10 (12): e0146028.
doi: 10.1371/journal.pone.0146028 |
| 32 |
BAO T T , YAO J W , ZHOU S , et al. Naringin prevents follicular atresia by inhibiting oxidative stress in the aging chicken[J]. Poult Sci, 2022, 101 (7): 101891.
doi: 10.1016/j.psj.2022.101891 |
| 33 |
JIANG J G , CHEN C L , CARD J W , et al. Cytochrome P450 2J2 promotes the neoplastic phenotype of carcinoma cells and is up-regulated in human tumors[J]. Cancer Res, 2005, 65 (11): 4707- 4715.
doi: 10.1158/0008-5472.CAN-04-4173 |
| 34 | GRESHAM R C H , BAHNEY C S , LEACH J K . Growth factor delivery using extracellular matrix-mimicking substrates for musculoskeletal tissue engineering and repair[J]. Bioact Mater, 2021, 6 (7): 1945- 1956. |
| 35 |
NY T , WAHLBERG P , BRÄNDSTRÖM I J M . Matrix remodeling in the ovary: regulation and functional role of the plasminogen activator and matrix metalloproteinase systems[J]. Mol Cell Endocrinol, 2002, 187 (1-2): 29- 38.
doi: 10.1016/S0303-7207(01)00711-0 |
| 36 | HRABIA A , SOCHA J K , SECHMAN A . Involvement of matrix metalloproteinases (MMP-2, -7, -9) and their tissue inhibitors (TIMP-2, -3) in the regression of chicken postovulatory follicles[J]. Gen Comp Endocrinol, 2018, 260, 32- 40. |
| 37 | NIKOLOUDAKI G . Functions of matricellular proteins in dental tissues and their emerging roles in orofacial tissue development, maintenance, and disease[J]. Int J Mol Sci, 2021, 22 (12): 6626. |
| 38 | GIBLIN S P , MIDWOOD K S . Tenascin-C: form versus function[J]. Cell Adh Migr, 2015, 9 (1-2): 48- 82. |
| 39 | CHIOVARO F , CHIQUET-EHRISMANN R , CHIQUET M . Transcriptional regulation of tenascin genes[J]. Cell Adh Migr, 2015, 9 (1-2): 34- 47. |
| 40 | GOOSSENS K , VAN SOOM A , VAN ZEVEREN A , et al. Quantification of fibronectin 1 (FN1) splice variants, including two novel ones, and analysis of integrins as candidate FN1 receptors in bovine preimplantation embryos[J]. BMC Dev Biol, 2009, 9, 1. |
| 41 | CUI Z F , LIU L B , KWAME AMEVOR F , et al. High expression of miR-204 in chicken atrophic ovaries promotes granulosa cell apoptosis and inhibits autophagy[J]. Front Cell Dev Biol, 2020, 8, 580072. |
| 42 | LI X K . The FGF metabolic axis[J]. Front Med, 2019, 13 (5): 511- 530. |
| 43 | PUSCHECK E E , PATEL Y , RAPPOLEE D A . Fibroblast growth factor receptor (FGFR)-4, but not FGFR-3 is expressed in the pregnant ovary[J]. Mol Cell Endocrinol, 1997, 132 (1-2): 169- 176. |
| 44 | ZHAO W J , ZHU L L , YANG W Q , et al. LPAR5 promotes thyroid carcinoma cell proliferation and migration by activating class IA PI3K catalytic subunit p110β[J]. Cancer Sci, 2021, 112 (4): 1624- 1632. |
| 45 | HERNÁNDEZ-BARRANCO A , SANTOS V , MAZARIEGOS M S , et al. NGFR regulates stromal cell activation in germinal centers[J]. Cell Rep, 2024, 43 (2): 113705. |
| [1] | Xiaoxu ZHANG, Hao LI, Pingjie FENG, Hao YANG, Xinyue LI, Ran LÜ, Zhangyuan PAN, Mingxing CHU. Application of Single-Cell Transcriptome Sequencing Technology in Domesticated Animals [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(8): 3276-3287. |
| [2] | Mingmin ZENG, Jun MENG, Yaqi ZENG, Jianwen WANG, Haifeng DENG, Wanlu REN, Yuheng XUE, Tingting SHANG, Feng GAO, Xinkui YAO. Analysis of Serum Proteomics in Early Pregnancy of Yili Horses Based on 4D-DIA Technology [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(8): 3493-3502. |
| [3] | Jing CHEN, Xuebei WU, Dongzhi MIAO, Chi ZHANG, Zhenyu GUO, Ying WANG. Comparative Analysis of Transcriptome of Pigeon Follicles at Early Stage of Laying Interval Reveals Genes Related to Follicular Development [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(8): 3503-3515. |
| [4] | Yaxuan MENG, Yan LIU, Jing WANG, Guoshun CHEN, Tao FENG. Research Progress in the Effect of Oxidative Stress on Ovarian Function in Female Livestock [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(7): 2825-2835. |
| [5] | Wanqing LI, Yaqi ZENG, Xinkui YAO, Jianwen WANG, Xinxin YUAN, Chen MENG, Yuanfang SUN, Xuan PENG, Jun MENG. Comparative Analysis of Blood Transcriptome in Yili Horses Bred for Meat Performance [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(7): 2951-2962. |
| [6] | Mingliang HE, Xiaoyang LÜ, Yongqing JIANG, Zhenghai SONG, Yeqing WANG, Huiguo YANG, Shanhe WANG, Wei SUN. Function Analysis of SOX18 in Hu Sheep Hair Follicle Dermal Papilla Cells Based on Transcriptome Sequencing [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(6): 2409-2420. |
| [7] | CHEN Zhe, QU Xiaolu, GUO Binbin, SUN Xuefeng, YAN Leyan. Study on Candidate Genes for Green Light Affecting Early Development of Goose Embryo Heart Based on Transcriptome Sequencing [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(5): 1978-1988. |
| [8] | XU Junjie, ZHANG Lutong, WANG Jinjie, CHEN Xiaochen, HE Weixian, CAI Chuanjiang, CHU Guiyan, YANG Gongshe. Exploring the Effect of Epimedium on Estrus of Gilts Based on Multiomics and Network Pharmacology [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(4): 1615-1628. |
| [9] | CAO Jinkang, ZHANG Chun, WANG Jiayao, LI Xiaotong, WANG Pengyu, FANG Yingyan, ZHANG Yu, DING Ning, JIANG Li. Proteomic Analysis of Sperm with Different Freezability in Chinese Holstein Bulls [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(3): 1052-1061. |
| [10] | WANG Xin, NIE Tong, LI Aqun, MA Jun. Hesperidin Alleviates High-fat-diet Induced Hepatic Oxidative Stress in Mice via Oxidative Phosphorylation Pathway [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(3): 1302-1313. |
| [11] | GAO Yawei, PENG Di, SUN Zhaoyang, YAN Ziyue, CUI Kai, MA Zefang. Mining the Molecular Mechanism of Exogenous Melatonin Affecting the Development of Mink Ovary Based on Transcriptome Data [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(2): 607-618. |
| [12] | Congying YU, Jinhua WU, Bingzhou ZHONG, Haiquan ZHAO, Shuwen TAN, Hui YU, Hua LI. Analysis of Whole Transcriptome Characteristics of the Hermaphroditic Pig's Pituitary Gland [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(11): 4925-4937. |
| [13] | ZHOU Hongtai, YAN Junrong, LI Pengfei. Transcriptome Sequencing was Used to Screen Genes Related to Follicular Development Bias and Dominant Follicle Selection in Cattle [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(11): 5059-5071. |
| [14] | LU Jian, JU Xiaojun, WANG Xingguo, MA Meng, WANG Qiang, LI Yongfeng, DOU Taocun, HU Yuping, GUO Jun, SHAO Dan, TONG Haibing, QU Liang. Effects of Metabolizable Energy Intake during Rearing on Development of Reproductive Organs, Hormone Level and Gene Expression in Ovary of Laying Hens [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(11): 5085-5100. |
| [15] | Tingting ZHOU, Li LI, Yantao WU, Wenying LU, Baoquan FU, Hong YIN, Wanzhong JIA, Hongbin YAN. Progress on Single-cell Transcriptomics Technology and Its Applications in Research on Parasites [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(10): 4290-4301. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||