





Acta Veterinaria et Zootechnica Sinica ›› 2025, Vol. 56 ›› Issue (11): 5433-5448.doi: 10.11843/j.issn.0366-6964.2025.11.008
• Animal Genetics and Breeding • Previous Articles Next Articles
JIANG Yifan1( ), LI Haoxing2,3,4(
), LI Haoxing2,3,4( ), LIN Yu1, ZHANG Zhihua2,3,4, WANG Yanfang1,*(
), LIN Yu1, ZHANG Zhihua2,3,4, WANG Yanfang1,*( )
)
Received:2025-04-08
Online:2025-11-23
Published:2025-11-27
Contact:
WANG Yanfang
E-mail:jiang092122@163.com;1076599497@qq.com;wangyanfang@caas.cn
CLC Number:
JIANG Yifan, LI Haoxing, LIN Yu, ZHANG Zhihua, WANG Yanfang. Three-Dimensional Genome Architecture of Porcine White and Beige Adipocytes[J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(11): 5433-5448.
Table 1
Primer sequences of qPCR"
| 基因 Gene | 上游引物(5′→3′) Forward primer | 下游引物(5′→3′) Reverse primer |
| 18S rRNA | GTAACCCGTTGAACCCCATT | CCATCCAATCGGTAGTAGCG |
| FABP4 | GGCCAAACCCAACCTGATCA | CATCCCACTTCTGCACCTGT |
| PPARg | GTCATGGGTGAAACTCTGGGA | TGTCAACCATGGTCACCTCTTG |
| UCP3 | CAACAGGAAGTACAGCGGGA | GTGATGTTGGGCAGAATTCCTTT |
| CIDEA | GTCAAGGCCACCATGTACGA | AGCATTCGGAGCATGTACGT |
| PDK4 | GGCAGCAGTGGTCCAAGAT | CGACTGTAGCCCTCATTGCA |
| LEPTIN | TGGCCCTATCTGTCCTACGT | TGGACAGACTCAGGACAGGA |
| ADCY5 | CAGACATCAACGCCAAGCAG | AAGCCCTCGATGTCAGCAAA |
| TMEM100 | ACCTGTGCTGTGTTGTGTTTG | TATCGCCCATTCACTGCACA |
| AMCF-Ⅱ | AGGCGGCTGTAGTGAGAGA | CCATTCTTCAGGGTGGCTATCA |
Table 2
Primers for functional elements in dual-luciferase assay and their genomic locations"
| 功能元件名称及基因组定位 Name and genomic localization of functional elements | 上游引物(5′→3′) Forward primer | 下游引物(5′→3′) Reverse primer |
| TMEM100 Promoter (chr12:3212541-32016094) | CTATCTGATGCCCTAGACCT | CAAATTGTAAATCAACTATACTCC |
| TMEM100 Enhancer 200 bp (chr12:31945275-31945473) | GGGAAGAAGACAGGGTGGAA | CTGTGTCTTAGTTTATTGCCAT |
| TMEM100 Enhancer 525 bp (chr12:31945100-31945623) | GCCTCCAAATCTTCTTTTGAG | TGGGAGTCACCAGTGTGGG |
| AMCF-Ⅱ Promoter (chr8:70006225-70008224) | TTGGTGCCCAAACTAATTCA | AAGGCGCGGAGATTTGAAG |
| AMCF-Ⅱ Enhancer 250 bp (chr8:70102263-70102550) | GACTGCACTTTCTTAGGTTTA | TTGTTTTAGCCAAAGGAATCT |
| AMCF-Ⅱ Enhancer 500 bp (chr8:70102150-70102650) | TTTTCCATCTGTAATGGAGCC | ACCTAGGGATTTAGCACAGG |
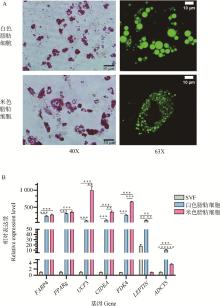
Fig. 1
Lipid droplet characteristics and mRNA expression levels in white and beige adipocytes A. Oil Red O staining (40×) and BODIPY 493/503 fluorescence staining (63×) of white and beige adipocytes. B. Relative expression levels of known adipocyte marker genes in SVF, white and beige adipocytes. All data is presented as the "mean±SEM", *.P < 0.05, **.P < 0.01, ***.P < 0.001, the same as below"

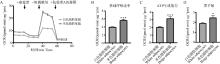
Fig. 2
Mitochondrial stress test in white and beige adipocytes A. Comparison of oxygen consumption rate (OCR) between white and beige adipocytes derived from SVF after 8-day differentiation; B-D. The basal respiration rate (B), ATP production (C) and proton leak (D) were calculated based on the OCRs"
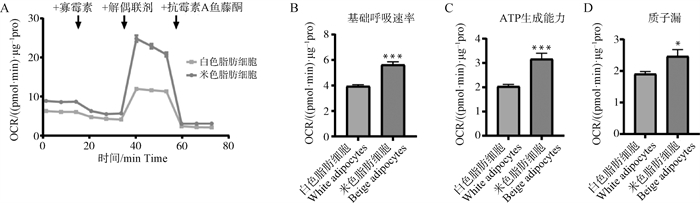
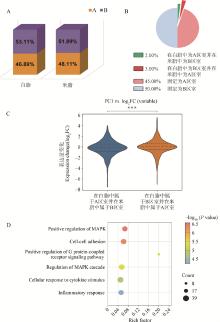
Fig. 4
The characteristics of compartment A/B between white and beige adipocytes A. Percentage of compartment A/B in white and beige adipocytes; B. Proportion of chromatin compartments in white versus beige adipocytes; C. Gene expression levels in white versus beige adipocytes across chromatin compartments; D. Functional enrichment analysis of genes transitioning from compartment B in white adipocytes to compartment A in beige adipocytes. P values were determined by Mann-Whitney U test"
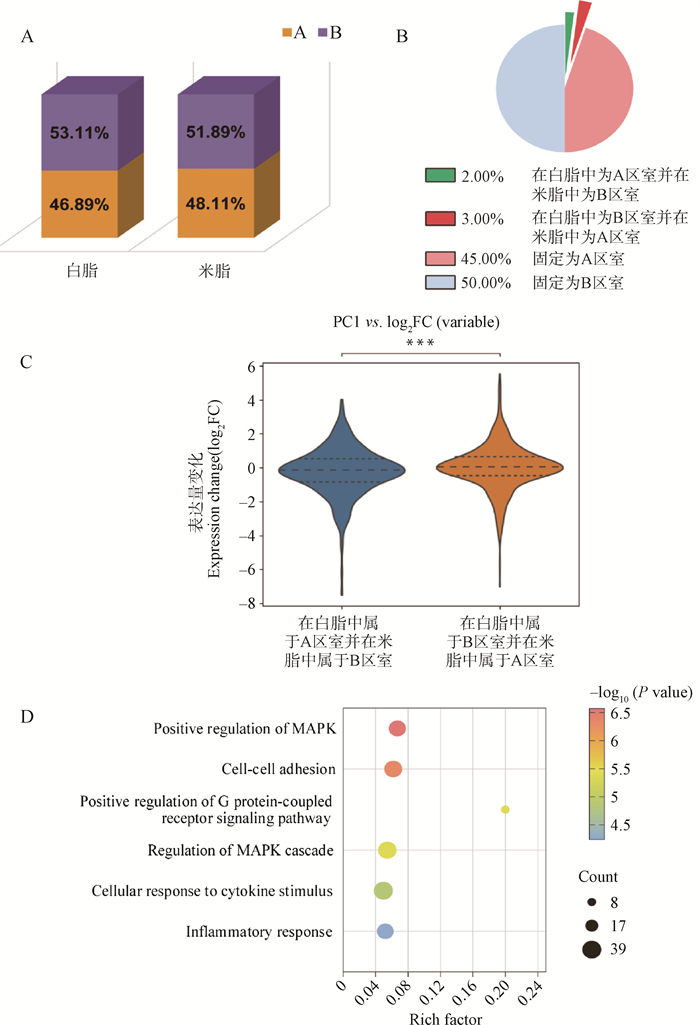
Table 3
Genes residing in B compartment in white adipocytes but A compartment in beige adipocytes, with specific up-regulation in the latter"
| 基因ID Gene ID | 基因名称 Gene name | 差异倍数 log2FC | 基因ID Gene ID | 基因名称 Gene name | 差异倍数 log2FC | |
| ENSSSCG00000009169 | SLC39A8 | 5.523 | ENSSSCG00000009120 | ZGRF1 | 2.249 | |
| ENSSSCG00000004597 | AQP9 | 4.990 | ENSSSCG00000007603 | NPTX2 | 2.209 | |
| ENSSSCG00000012129 | GLRA2 | 4.873 | ENSSSCG00000060144 | CCNA2 | 2.146 | |
| ENSSSCG00000060220 | NA | 4.625 | ENSSSCG00000011624 | MCM2 | 2.111 | |
| ENSSSCG00000059641 | NA | 4.278 | ENSSSCG00000003888 | STIL | 2.105 | |
| ENSSSCG00000009543 | MYO16 | 4.259 | ENSSSCG00000012150 | REPS2 | 2.073 | |
| ENSSSCG00000053166 | NA | 3.858 | ENSSSCG00000039760 | PRR16 | 2.046 | |
| ENSSSCG00000037821 | RGS5 | 3.823 | ENSSSCG00000059773 | NA | 2.043 | |
| ENSSSCG00000053781 | NA | 3.376 | ENSSSCG00000004157 | IL20RA | 2.007 | |
| ENSSSCG00000025240 | DDIT4L | 3.353 | ENSSSCG00000035355 | F2R | 1.895 | |
| ENSSSCG00000013273 | CHST1 | 3.052 | ENSSSCG00000009446 | PCDH17 | 1.779 | |
| ENSSSCG00000012540 | SLC25A53 | 2.898 | ENSSSCG00000015959 | RAPGEF4 | 1.770 | |
| ENSSSCG00000015961 | CDCA7 | 2.726 | ENSSSCG00000041777 | K7GN43 | 1.734 | |
| ENSSSCG00000030016 | PDE9A | 2.655 | ENSSSCG00000056445 | NA | 1.715 | |
| ENSSSCG00000026527 | HCN1 | 2.471 | ENSSSCG00000010647 | ADRB1 | 1.706 | |
| ENSSSCG00000025965 | SPDL1 | 2.356 | ENSSSCG00000039027 | TRO | 1.701 | |
| ENSSSCG00000024325 | SGK2 | 2.323 | ENSSSCG00000023434 | PPM1L | 1.541 | |
| ENSSSCG00000015136 | UBASH3B | 2.294 | ENSSSCG00000017254 | MAP2K6 | 1.517 | |
| ENSSSCG00000034229 | MIS18A | 2.274 | ENSSSCG00000010912 | KIF14 | 1.509 |
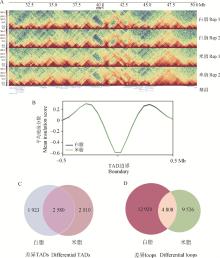
Fig. 6
Differential TADs and chromatin loops between white and beige adipocytes A.Normalized Hi-C contact heatmap of 20 Mb region in chromosome 1 were showed at 50 kb resolution in white and beige adipocytes; B. Mean value of insulation scores around white and beige adipocytes TAD boundary; C. Differential TADs between white and beige adipocytes; D. Differential loops between white and beige adipocytes"

| 1 |
LIN J , CAO C , TAO C , et al. Cold adaptation in pigs depends on UCP3 in beige adipocytes[J]. J Mol Cell Biol, 2017, 9 (5): 364- 375.
doi: 10.1093/jmcb/mjx018 |
| 2 |
WU J , BOSTRöM P , SPARKS L M , et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human[J]. Cell, 2012, 150 (2): 366- 376.
doi: 10.1016/j.cell.2012.05.016 |
| 3 |
XUE R , LYNES M D , DREYFUSS J M , et al. Clonal analyses and gene profiling identify genetic biomarkers of the thermogenic potential of human brown and white preadipocytes[J]. Nat Med, 2015, 21 (7): 760- 768.
doi: 10.1038/nm.3881 |
| 4 |
SHINODA K , LUIJTEN I H , HASEGAWA Y , et al. Genetic and functional characterization of clonally derived adult human brown adipocytes[J]. Nat Med, 2015, 21 (4): 389- 394.
doi: 10.1038/nm.3819 |
| 5 |
ZHANG L , HU S , CAO C , et al. Functional and genetic characterization of porcine beige adipocytes[J]. Cells, 2022, 11 (4): 751.
doi: 10.3390/cells11040751 |
| 6 |
LI F , WANG D , SONG R , et al. The asynchronous establishment of chromatin 3D architecture between in vitro fertilized and uniparental preimplantation pig embryos[J]. Genome Biol, 2020, 21 (1): 203.
doi: 10.1186/s13059-020-02095-z |
| 7 |
ZHAO Y , HOU Y , XU Y , et al. A compendium and comparative epigenomics analysis of cis-regulatory elements in the pig genome[J]. Nat Commun, 2021, 12 (1): 2217.
doi: 10.1038/s41467-021-22448-x |
| 8 |
YU L , HUANG T , LIU S , et al. The landscape of super-enhancer regulates remote target gene transcription through loop domains in adipose tissue of pig[J]. Heliyon, 2024, 10 (4): e25725.
doi: 10.1016/j.heliyon.2024.e25725 |
| 9 |
ZHAO Y , LI X , YU W , et al. Differential expression of ADRB1 causes different responses to norepinephrine in adipocytes of Duroc-Landrace-Yorkshire pigs and min pigs[J]. J Therm Biol, 2024, 123, 103906.
doi: 10.1016/j.jtherbio.2024.103906 |
| 10 |
XUAN M F , LUO Z B , HAN S Z , et al. Skeletal muscle-secreted myokine interleukin-6 induces white adipose tissue conversion into beige adipose tissue in myostatin gene knockout pigs[J]. Domest Anim Endocrinol, 2022, 78, 106679.
doi: 10.1016/j.domaniend.2021.106679 |
| 11 |
ZHAO Y , LI X , TIAN Y , et al. Nonivamide induces brown fat-like characteristics in porcine subcutaneous adipocytes[J]. Biochem Biophys Res Commun, 2022, 619, 68- 75.
doi: 10.1016/j.bbrc.2022.06.047 |
| 12 |
LIU J , JIANG Y , CHEN C , et al. Bone morphogenetic protein 2 enhances porcine beige adipogenesis via AKT/mTOR and MAPK signaling pathways[J]. Int J Mol Sci, 2024, 25 (7): 3915.
doi: 10.3390/ijms25073915 |
| 13 | KIM S , YAZAWA T , KOIDE A , et al. Potential role of pig UCP3 in modulating adipocyte browning via the beta-adrenergic receptor signaling pathway[J]. Biology (Basel), 2024, 13 (5): 284. |
| 14 |
SPITZ F , FURLONG E E . Transcription factors: from enhancer binding to developmental control[J]. Nat Rev Genet, 2012, 13 (9): 613- 626.
doi: 10.1038/nrg3207 |
| 15 |
LEVINE M , CATTOGLIO C , TJIAN R . Looping back to leap forward: transcription enters a new era[J]. Cell, 2014, 157 (1): 13- 25.
doi: 10.1016/j.cell.2014.02.009 |
| 16 |
BONEV B , CAVALLI G . Organization and function of the 3D genome[J]. Nat Rev Genet, 2016, 17 (11): 661- 678.
doi: 10.1038/nrg.2016.112 |
| 17 | 陈川河, 刘嘉莉, 张立兰, 等. 猪SGK家族基因的生物信息学分析及其在猪脂肪组织和细胞中的表达[J]. 中国畜牧兽医, 2022, 49 (9): 3310- 3320. |
| CHEN C H , LIU J L , ZHANG L L , et al. Bioinformatics analysis of porcine SGK family genes and their expression in porcine adipose tissues and adipocytes[J]. China Animal Husbandry & Veterinary Medicine, 2022, 49 (9): 3310- 3320. | |
| 18 |
RAO S S , HUNTLEY M H , DURAND N C , et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping[J]. Cell, 2014, 159 (7): 1665- 1680.
doi: 10.1016/j.cell.2014.11.021 |
| 19 |
UETA C B , FERNANDES G W , CAPELO L P , et al. β(1) Adrenergic receptor is key to cold-and diet-induced thermogenesis in mice[J]. J Endocrinol, 2012, 214 (3): 359- 365.
doi: 10.1530/JOE-12-0155 |
| 20 | IYER M S , PASZKIEWICZ R L , BERGMAN R N , et al. Activation of NPRs and UCP1-independent pathway following CB1R antagonist treatment is associated with adipose tissue beiging in fat-fed male dogs[J]. Am J Physiol Endocrinol Metab, 2019, 317 (3): E535-e47. |
| 21 |
CEDDIA R P , LIU D , SHI F , et al. Increased energy expenditure and protection from diet-induced obesity in mice lacking the cGMP-specific phosphodiesterase PDE9[J]. Diabetes, 2021, 70 (12): 2823- 2836.
doi: 10.2337/db21-0100 |
| 22 |
WANG B , ZHOU Q , BI Y , et al. Phosphatase PPM1L prevents excessive inflammatory responses and cardiac dysfunction after myocardial infarction by inhibiting IKKβ activation[J]. J Immunol, 2019, 203 (5): 1338- 1347.
doi: 10.4049/jimmunol.1900148 |
| 23 |
CAO W , HUANG H , XIA T , et al. Homeobox a5 promotes white adipose tissue browning through inhibition of the tenascin C/Toll-like receptor 4/nuclear factor kappa B inflammatory signaling in mice[J]. Front Immunol, 2018, 9, 647.
doi: 10.3389/fimmu.2018.00647 |
| 24 |
HE M , LI Y , TANG Q , et al. Genome-wide chromatin structure changes during adipogenesis and myogenesis[J]. Int J Biol Sci, 2018, 14 (11): 1571- 1585.
doi: 10.7150/ijbs.25328 |
| 25 |
DEKKER J , MARTI-RENOM M A , MIRNY L A . Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data[J]. Nat Rev Genet, 2013, 14 (6): 390- 403.
doi: 10.1038/nrg3454 |
| 26 |
WHYTE W A , ORLANDO D A , HNISZ D , et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes[J]. Cell, 2013, 153 (2): 307- 319.
doi: 10.1016/j.cell.2013.03.035 |
| 27 | 王彦芳, 姜琳, 陶聪, 等. TMEM100基因调控猪脂肪细胞能量代谢: 中国, 202311401055.9[P]. 2024-02-13. |
| WANG Y F, JIANG L, TAO C, et al. The TMEM100 gene regulates energy metabolism in porcine adipocytes: China, 202311401055.9[P]. 2024-02-13. (in Chinese) | |
| 28 |
SEPURU K M , POLURI K M , RAJARATHNAM K . Solution structure of CXCL5--a novel chemokine and adipokine implicated in inflammation and obesity[J]. PLoS One, 2014, 9 (4): e93228.
doi: 10.1371/journal.pone.0093228 |
| 29 |
ALHAMOUD Y , WU J , AHMAD M I , et al. 6-Gingerol induces browning of white adipose tissue to improve obesity through PI3K/AKT-RPS6-mediated thermogenesis pathway[J]. Food Frontiers, 2023, 4 (3): 1285- 1297.
doi: 10.1002/fft2.251 |
| 30 |
LEE H J , LEE J , YANG M J , et al. Endothelial cell-derived stem cell factor promotes lipid accumulation through c-Kit-mediated increase of lipogenic enzymes in brown adipocytes[J]. Nat Commun, 2023, 14 (1): 2754.
doi: 10.1038/s41467-023-38433-5 |
| 31 |
FULCO C P , MUNSCHAUER M , ANYOHA R , et al. Systematic mapping of functional enhancer-promoter connections with CRISPR interference[J]. Science, 2016, 354 (6313): 769- 773.
doi: 10.1126/science.aag2445 |
| [1] | LIN Xiao, LI Ruijie, LIU Long, GENG Tuoyu, GONG Daoqing. Research Progress on Sex Determining Genes and Their Methylation Regulation in Animals [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(9): 4129-4142. |
| [2] | TIAN Jiao, LONG Juyan, CHEN Xia, CEN Xiaoli, NIU Xi, HUANG Shihui, WANG Jiafu, RAN Xueqin. Down-regulation of Gene Expression by SINE Insertion in the 3'UTR of the ENTPD1 Gene in the Xiang Pig [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(9): 4303-4314. |
| [3] | QIN Yang, XIA Siting, HE Liuqin, WANG Tianli, LIU Yuyan, JIANG Xiaohan, LIU Zhihao, LIU Siwei, LI Tiejun, YIN Yulong. Effect of Chronic Oxidative Stress on Trace Elements in Organ Tissues of Weaned Piglets [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(9): 4452-4460. |
| [4] | RU Min, JIANG Xiaofeng, LUO Guosheng, WU Yonghou. Effects of Dietary Bacillus subtilis Supplementation on Growth Performance, Serum Immunity and Antioxidant Function, Intestinal Morphology and Microorganisms of Piglets Challenged with Escherichia coli [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(9): 4461-4471. |
| [5] | LIU Can, SU Yixin, JING Xianjin, LI Wenze, YANG Lepu, WANG Ruijun, ZHANG Yanjun, WANG Zhiying, LÜ Qi, SU Rui. Research Progress of Epigenetics in Sheep and Goats Genetic Breeding [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(8): 3561-3577. |
| [6] | YU Shulong, MAO Nannan, WANG Yunlong, ZHANG Yiran, WANG Yuanyuan, ZHOU Rongyan, ZANG Sumin, XIE Hui. Research Progress on Pigeon Sex Identification [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(8): 3601-3609. |
| [7] | CHI Shunshun, WU Dan, WANG Nan, WANG Wanjie, NIE Yuxin, MU Yulian, LIU Zhiguo, ZHU Zhendong, LI Kui. Establishment and Application of A Detection Method for MSTN Gene-Edited Pigs Based on RPA-CRISPR/Cas12a [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(8): 3734-3748. |
| [8] | LIU Sha, YANG Caichun, ZHANG Xiaoyu, CHEN Qiong, LIU Xiong, CHEN Hongbo, ZHOU Huanhuan, SHI Liangyu. Population Genetic Structure and Genome-wide Runs of Homozygosity Analysis in Meihuaxing Pigs Based on 80K SNP Chip [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(8): 3749-3760. |
| [9] | CAO Ning, ZHANG Hu, WANG Junli, SA Renna, ZHAO Feng, XIE Jingjing, GAO Lixiang, ZHAO Jiangtao, DONG Ying, WANG Yuming. Effect of Drying Method on Determination of Amino Acid Digestibility of Pig Feed by Biomimetic Method [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(8): 3893-3907. |
| [10] | MENG Yaxuan, LIU Yan, WANG Jing, CHEN Guoshun, FENG Tao. Effects of Glucosamine on Serum Anti-oxidation, Inflammatory Indexes and Intestinal Microbes in Weaned Piglets [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(8): 3908-3921. |
| [11] | WANG Nan, WANG Chengming, WANG Jing, LIN Xingtong, HE Lingyun. Effects of Phosphatidylethanolamine on Colonic Mucosal Barrier Function and Gut Microbiota in Postnatal Growth Retardation Piglets [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(7): 3305-3315. |
| [12] | SHU Jingchao, ZHANG Han, PENG Zhifeng, QIAO Hongxing. Isolation, Identification and Virulence Gene Analysis of Porcine Pathogenic Lactococcus garvieae [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(7): 3399-3407. |
| [13] | ZHAO Shunran, FU Guixin, PANG Zhaoqi, XIA Wei, LI Junjie, TAO Chenyu. Research Progress on the Mechanism of Porcine Granulosa Cells in Follicular Atresia [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(6): 2537-2545. |
| [14] | WU Tong, WANG Nan, XING Yuxin, ZHANG Ben, HU Panyang, ZHANG Haitao, ZHU Yufeng, WU Xiangzhe, YANG Feng, LI Xiuling, WANG Kejun, HAN Xuelei, LI Xinjian, YU Tong, BAI Jun, LI Gaiying, QIAO Ruimin. Association Analysis of Backfat Thickness and Genome-Wide Copy Number Variations in Yunan Black Pigs [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(6): 2639-2648. |
| [15] | LIU Siqi, YANG Zhen, YANG Yanan, CAI Yuan, ZHAO Shengguo. The Effect of Interfering with AdiopR2 on the Thermogenesis of Subcutaneous Inguinal Adipocytes in Tibetan Pigs [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(6): 2649-2660. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||