





Acta Veterinaria et Zootechnica Sinica ›› 2025, Vol. 56 ›› Issue (6): 2857-2867.doi: 10.11843/j.issn.0366-6964.2025.06.029
• Preventive Veterinary Medicine • Previous Articles Next Articles
WANG Yan1( ), HUANG Xinyu1, WU Guiying1, WU Wanping1, Lü Qizhuang1,2,*(
), HUANG Xinyu1, WU Guiying1, WU Wanping1, Lü Qizhuang1,2,*( )
)
Received:2024-06-08
Online:2025-06-23
Published:2025-06-25
Contact:
Lü Qizhuang
E-mail:wy2765770090@163.com;lvqizhuang062@163.com
CLC Number:
WANG Yan, HUANG Xinyu, WU Guiying, WU Wanping, Lü Qizhuang. Genetic Variation Analysis of Porcine Circovirus Type 3 in China from 2017 to 2023[J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(6): 2857-2867.
Table 1
Detection results of PCV3 gene subtypes in China from 2017 to 2023"
| 年份 Year | 不同亚型检测率/% Detection rate of different gene subtype | |||||||
| 3a-1 | 3a-2 | 3a-3 | 3b-1 | 3b-2 | 3b-3 | 3b-4 | 3b-5 | |
| 2017 | 12.31 | 30.77 | 1.54 | 1.54 | 1.54 | 30.77 | 6.15 | 15.38 |
| 2018 | 21.95 | 31.71 | 2.44 | 4.88 | 4.88 | 17.07 | 9.76 | 9.76 |
| 2019 | 23.53 | 41.18 | 5.88 | 0.00 | 17.65 | 0.00 | 0.00 | 11.76 |
| 2020 | 23.08 | 7.70 | 0.00 | 0.00 | 69.23 | 0.00 | 0.00 | 0.00 |
| 2021 | 12.50 | 25.00 | 0.00 | 12.50 | 0.00 | 25.00 | 0.00 | 25.00 |
| 2022 | 0.00 | 0.00 | 0.00 | 0.00 | 20.00 | 0.00 | 20.00 | 60.00 |
| 2023 | 73.68 | 0.00 | 0.00 | 0.00 | 0.00 | 21.05 | 0.00 | 5.26 |
| 总计 Total | 22.62 | 25.60 | 1.79 | 23.81 | 9.52 | 19.64 | 5.36 | 13.10 |
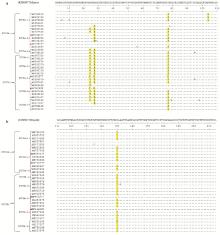
Fig. 4
Comparative analysis of amino acid sequence of PCV3 Cap protein a. Comparative analysis of amino acid sequences at positions 1-110; b. Comparative analysis of amino acid sequences at positions 111-214. KT869077 is the standard sequence, and the four amino acid sequences marked with the five-pointed star are the representative sequences screened in this study. If the residues are completely consistent, they are indicated by "·""
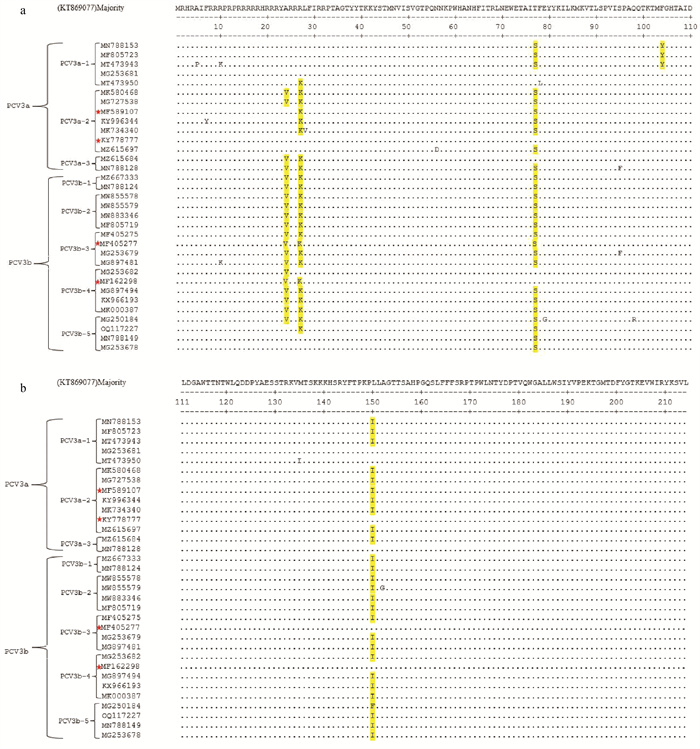

Fig. 5
Prediction results of secondary structure, surface position and B cell epitope of amino acid sequence of some PCV3 Cap proteins E represents the epitope, orange represents the predicted protein sequence of BepiPred-2.0, H-pink, E-blue and C-orange represent the heldicted relative surface accessibility. The figure shows eight representative amino acid sequences of PCV3 Cap protein"
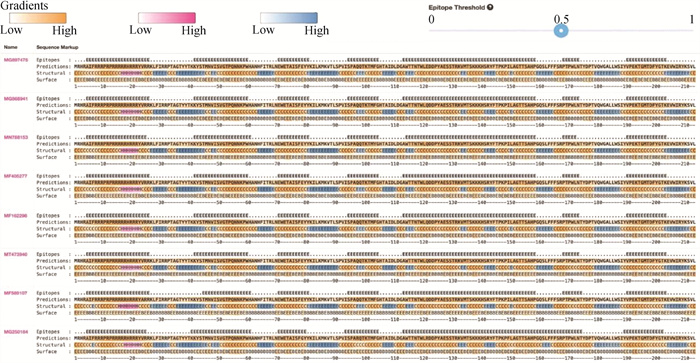

Fig. 6
Identification of recombination events MN788136 is the only recombinant, which is between MN788135 and another unknown sequence. The window size is 30, the y axis represents the base position, the x axis represents the position of the information site, and the left and right boundaries of the pink part represent the breakpoint position"
| 1 |
韩阳, 关帅印, 李振, 等. 猪圆环病毒3型Rep蛋白的原核表达及酶活性分析[J]. 畜牧兽医学报, 2024, 55 (5): 2061- 2071.
doi: 10.11843/j.issn.0366-6964.2024.05.024 |
|
HAN Y , GUANG S Y , LI Z , et al. Prokaryotic expression and protein activity of porcine circovirus type 3 Rep[J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55 (5): 2061- 2071.
doi: 10.11843/j.issn.0366-6964.2024.05.024 |
|
| 2 | TISCHER I , RASCH R , TOCHTERMANN G . Characterization of papovavirus-and picornavirus-like particles in permanent pig kidney cell lines[J]. Zentralbl Bakteriol Orig A, 1974, 226 (2): 153- 167. |
| 3 |
ALLAN G M , MC NEILLY F , MEEHAN B M , et al. Isolation and characterisation of circoviruses from pigs with wasting syndromes in Spain, Denmark and Northern Ireland[J]. Vet Microbiol, 1999, 66 (2): 115- 123.
doi: 10.1016/S0378-1135(99)00004-8 |
| 4 |
RODRIGUES I L F , CRUZ A C M , SOUZA A E , et al. Retrospective study of porcine circovirus 3 (PCV3) in swine tissue from Brazil (1967-2018)[J]. Braz J Microbiol, 2020, 51 (3): 1391- 1397.
doi: 10.1007/s42770-020-00281-6 |
| 5 |
YANG Y T , XU T , WEN J H , et al. Prevalence and phylogenetic analysis of porcine circovirus type 2 (PCV2) and type 3 (PCV3) in the Southwest of China during 2020-2022[J]. Front Vet Sci, 2022, 9, 1042792.
doi: 10.3389/fvets.2022.1042792 |
| 6 | PALINSKI R , PIÑEYRO P , SHANG P , et al. A novel porcine circovirus distantly related to known circoviruses is associated with porcine dermatitis and nephropathy syndrome and reproductive failure[J]. J Virol, 2016, 91 (1): e01879- 16. |
| 7 |
ZHANG H H , HU W Q , LI J Y , et al. Novel circovirus species identified in farmed pigs designated as Porcine circovirus 4, Hunan province, China[J]. Transbound Emerg Dis, 2020, 67 (3): 1057- 1061.
doi: 10.1111/tbed.13446 |
| 8 | NIU G Y , ZHANG X W , JI W L , et al. Porcine circovirus 4 rescued from an infectious clone is replicable and pathogenic in vivo[J]. Transbound Emerg Dis, 2022, 69 (5): e1632- e1641. |
| 9 |
XU P L , ZHANG Y , ZHAO Y , et al. Detection and phylogenetic analysis of porcine circovirus type 3 in central China[J]. Transbound Emerg Dis, 2018, 65 (5): 1163- 1169.
doi: 10.1111/tbed.12920 |
| 10 |
KU X , CHEN F , LI P , et al. Identification and genetic characterization of porcine circovirus type 3 in China[J]. Transbound Emerg Dis, 2017, 64 (3): 703- 708.
doi: 10.1111/tbed.12638 |
| 11 |
SHEN H , LIU X , ZHANG P , et al. Genome characterization of a porcine circovirus type 3 in South China[J]. Transbound Emerg Dis, 2018, 65 (1): 264- 266.
doi: 10.1111/tbed.12639 |
| 12 |
HAYASHI S , OHSHIMA Y , FURUYA Y , et al. First detection of porcine circovirus type 3 in Japan[J]. J Vet Med Sci, 2018, 80 (9): 1468- 1472.
doi: 10.1292/jvms.18-0079 |
| 13 |
SUKMAK M , THANANTONG N , POOLPERM P , et al. The retrospective identification and molecular epidemiology of porcine circovirus type 3 (PCV3) in swine in Thailand from 2006 to 2017[J]. Transbound Emerg Dis, 2019, 66 (1): 611- 616.
doi: 10.1111/tbed.13057 |
| 14 |
FUX R , SÖCKLER C , LINK E K , et al. Full genome characterization of porcine circovirus type 3 isolates reveals the existence of two distinct groups of virus strains[J]. Virol J, 2018, 15 (1): 25.
doi: 10.1186/s12985-018-0929-3 |
| 15 |
KIM S C , NAZKI S , KWON S , et al. The prevalence and genetic characteristics of porcine circovirus type 2 and 3 in Korea[J]. BMC Vet Res, 2018, 14 (1): 294.
doi: 10.1186/s12917-018-1614-x |
| 16 |
SARAIVA G L , VIDIGAL P M P , ASSAO V S , et al. Retrospective detection and genetic characterization of porcine circovirus 3 (PCV3)strains identified between 2006 and 2007 in Brazil[J]. Viruses, 2019, 11 (3): 201.
doi: 10.3390/v11030201 |
| 17 | 曹旭阳, 袁红, 孙普, 等. 猪圆环病毒3型研究进展[J]. 中国兽医杂志, 2023, 59 (9): 1- 7. |
| CAO X Y , YUAN H , SUN P , et al. Research Progress on Porcine Circovirus Type 3[J]. Chinese Journal of Veterinary Medicine, 2023, 59 (9): 1- 7. | |
| 18 |
FU X , FANG B , MA J , et al. Insights into the epidemic characteristics and evolutionary history of the novel porcine circovirus type 3 in southern China[J]. Transbound Emerg Dis, 2018, 65 (2): e296- e303.
doi: 10.1111/tbed.12752 |
| 19 |
GE M , REN J , XIE Y L , et al. Prevalence and genetic analysis of porcine circovirus 3 in China from 2019 to 2020[J]. Front Vet Sci, 2021, 8, 773912.
doi: 10.3389/fvets.2021.773912 |
| 20 |
LI G , HE W , ZHU H , et al. Origin, genetic diversity, and evolutionary dynamics of novel porcine circovirus 3[J]. Adv Sci (Weinh), 2018, 5 (9): 1800275.
doi: 10.1002/advs.201800275 |
| 21 |
CHUNG H C , NGUYEN V G , PARK Y H , et al. Genotyping of PCV3 based on reassembled viral gene sequences[J]. Vet Med Sci, 2021, 7 (2): 474- 482.
doi: 10.1002/vms3.374 |
| 22 |
XIE J M , CHEN Y R , CAI G J , et al. Tree Visualization by one table (tvBOT): a web application for visualizing, modifying and annotating phylogenetic trees[J]. Nucleic Acids Res, 2023, 51 (W1): W587- W592.
doi: 10.1093/nar/gkad359 |
| 23 |
LV Q Z , WANG T , DENG J H , et al. Genomic analysis of porcine circovirus type 2 from southern China[J]. Vet Med Sci, 2020, 6 (4): 875- 889.
doi: 10.1002/vms3.288 |
| 24 |
FRANZO G , SEGALÉS J . Porcine circovirus 2 (PCV-2) genotype update and proposal of a new genotyping methodology[J]. PLoS One, 2018, 13 (12): e0208585.
doi: 10.1371/journal.pone.0208585 |
| 25 | 李周勉, 邹亚文, 蒋一凡, 等. 猪圆环病毒3型Cap蛋白序列的生物信息学研究[J]. 经济动物学报, 2020, 24 (1): 9- 16. |
| LI Z M , ZOU Y W , JIANG Y F , et al. Bioinformatics analysis of cap protein sequences of porcine circovirus type 3[J]. Journal of Economic Animal, 2020, 24 (1): 9- 16. | |
| 26 |
SAPORITI V , VALLS L , MALDONADO J , et al. Porcine circovirus 3 detection in aborted fetuses and stillborn piglets from swine reproductive failure cases[J]. Viruses, 2021, 13 (2): 264.
doi: 10.3390/v13020264 |
| 27 |
GRAU-ROMA L , STOCKMARR A , KRISTENSEN C S , et al. Infectious risk factors for individual postweaning multisystemic wasting syndrome (PMWS) development in pigs from affected farms in Spain and Denmark[J]. Res Vet Sci, 2012, 93 (3): 1231- 1240.
doi: 10.1016/j.rvsc.2012.07.001 |
| 28 |
ARRUDA B , PINEYRO P , DERSCHEID R , et al. PCV3-associated disease in the United States swine herd[J]. Emerg Microbes Infect, 2019, 8 (1): 684- 698.
doi: 10.1080/22221751.2019.1613176 |
| 29 |
QI S S , SU M J , GUO D H , et al. Molecular detection and phylogenetic analysis of porcine circovirus type 3 in 21 Provinces of China during 2015-2017[J]. Transbound Emerg Dis, 2019, 66 (2): 1004- 1015.
doi: 10.1111/tbed.13125 |
| 30 |
HA Z , LI J F , XIE C Z , et al. Prevalence, pathogenesis, and evolution of porcine circovirus type 3 in China from 2016 to 2019[J]. Vet Microbiol, 2020, 247, 108756.
doi: 10.1016/j.vetmic.2020.108756 |
| 31 |
SHEN H Q , LIU X H , ZHANG P F , et al. Porcine circovirus 3 Cap inhibits type I interferon signaling through interaction with STAT2[J]. Virus Res, 2020, 275, 197804.
doi: 10.1016/j.virusres.2019.197804 |
| 32 |
JIANG M , GUO J Q , ZHANG G P , et al. Fine mapping of linear B cell epitopes on capsid protein of porcine circovirus 3[J]. Appl Microbiol Biotechnol, 2020, 104 (14): 6223- 6234.
doi: 10.1007/s00253-020-10664-2 |
| [1] | WANG Shujuan, WANG Dongfang, MA Zhenyuan, ZHAO Xueli, LIU Ying, YANG Haibo, CHAI Mao, WANG Cui, YAN Ruoqian. Isolation, Identification and Evolutionary Analysis of a Strain of Senecavirus A [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(6): 3041-3046. |
| [2] | WANG Yanan, GUO Yaru, JIANG Yanping, CUI Wen, LI Jiaxuan, LI Yijing, WANG Li. Isolation, Identification and Pathogenicity Analysis of Porcine Rotavirus [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(5): 2259-2269. |
| [3] | LI Ting, ZHANG Chengcheng, WANG Xiuling, ZHANG Xiaorong, WU Yantao. Isolation and Identification of a Novel Duck Reovirus Strain and the Sequence Analysis of Its σC Gene [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(5): 2520-2524. |
| [4] | Bilin XIE, Zhimin LIN, Binbin LIN, Yijuan XU, Fengqiang LIN, Lu YAN, Huini WU, Cuiting LI, Haiou ZHOU, Zhaolong LI. Isolation, Identification and Pathogenicity Analysis of Riemerella anatipestifer Strain LC1 and CX1 [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(9): 4196-4203. |
| [5] | Yue LI, Changchun ZHANG, Guangyu LIU, Mengyuan GAO, Chaojun FU, Jiabao XING, Sijia XU, Qiyuan KUANG, Jing LIU, Xiaopeng GAO, Heng WANG, Lang GONG, Guihong ZHANG, Yankuo SUN. Application and Analysis of Meta-transcriptomics Sequencing Technology in the Diagnosis of Viral Diarrhea Diseases in Piglets [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(8): 3579-3589. |
| [6] | Haotian ZHU, Luyao LIU, Congshan YANG, Ji JIANG, Han SHI, Xiaoxiao LIU, Muxin CHEN, Qianming XU. Morphological Observation and Molecular Characterization of Anisakis Derived from Dolphin [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(6): 2599-2606. |
| [7] | HAN Yang, GUAN Shuaiyin, LI Zhen, ZHOU Saisai, YUAN Honggen, SONG Yunfeng. Prokaryotic Expression and Protein Activity of Porcine Circovirus Type 3 Rep [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(5): 2061-2071. |
| [8] | ZHANG Baoge, HUANG Yaqin, CAI Jinshuang, ZHU Chenguang, LI Yufeng. Preparation of Monoclonal Antibodies to Porcine Circovirus Type 3 Cap Protein and the Establishment of a Blocking ELISA Assay [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(3): 1170-1178. |
| [9] | JIANG Lingling, NIU Xiaoxia, LIU Qiang, ZHANG Gang, WANG Pu, LI Yong. Analysis of Infection Status of Beef Cattle Diarrhea-related Virus in Ningxia [J]. Acta Veterinaria et Zootechnica Sinica, 2023, 54(9): 3863-3871. |
| [10] | YANG Qing, GONG Jing, ZHAO Xueyan, ZHU Xiaodong, GENG Liying, ZHANG Chuansheng, WANG Jiying. Comparison of Array and Resequencing in Pig Genetic Structure Studies [J]. Acta Veterinaria et Zootechnica Sinica, 2023, 54(7): 2772-2782. |
| [11] | KU Xugang, YU Xuexiang, SUN Qi, LI Panpan, ZHANG Mengjia, LUO Rui, QIAN Ping, HE Qigai. Isolation and Pathogenicity Analysis of Porcine Circovirus Type 3 [J]. Acta Veterinaria et Zootechnica Sinica, 2023, 54(4): 1568-1578. |
| [12] | NI Zheng, YE Weicheng, CHEN Liu, YUN Tao, HUA Jionggang, ZHU Yinchu, ZHANG Cun. Genetic Variation and Pathogenicity of a Pseudorabies Virus Variant Strain in Mice [J]. Acta Veterinaria et Zootechnica Sinica, 2023, 54(4): 1766-1770. |
| [13] | LÜ Xuan, JIA Beiping, ZHU Meng, YU Shengzu, ZHANG Miao, WANG Bei, ZHU Yingqi, ZHANG Yunkai, WANG Qing, WANG Guijun. Genomic Characteristics and Pathogenicity Analysis of One Variant Strain of Goose Astrovirus [J]. Acta Veterinaria et Zootechnica Sinica, 2022, 53(8): 2812-2818. |
| [14] | HE Qing, LI Zhoumian, WANG Huan, LI Dantong, TAN Qinghui, ZOU Yawen, SU Jianming, DENG Zhibang, YANG Yi, WANG Naidong. Detection and Genetic Evolution of Porcine Circovirus Type 3 in Swine Aborted Fetus [J]. Acta Veterinaria et Zootechnica Sinica, 2022, 53(5): 1527-1535. |
| [15] | YOU Guishuang, YI Xingyou, JIANG Meishan, LI Zhongwei, TAN Helin, CAO Chi, SHU Gang, TIAN Yaofu, ZHU Qing, ZHAO Xiaoling. Study on the Genetic Relationship of Six Local Chicken Populations from Sichuan Basin [J]. Acta Veterinaria et Zootechnica Sinica, 2022, 53(2): 370-380. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||