





Acta Veterinaria et Zootechnica Sinica ›› 2025, Vol. 56 ›› Issue (6): 2661-2671.doi: 10.11843/j.issn.0366-6964.2025.06.012
• Animal Genetics and Breeding • Previous Articles Next Articles
LIU Sha1,2( ), SU Meng2, GAO Qianmei2, SONG Danli1,2, ZHAO Guiping2, LI Jianhui1,*(
), SU Meng2, GAO Qianmei2, SONG Danli1,2, ZHAO Guiping2, LI Jianhui1,*( ), LI Qinghe2,*(
), LI Qinghe2,*( )
)
Received:2024-11-11
Online:2025-06-23
Published:2025-06-25
Contact:
LI Jianhui, LI Qinghe
E-mail:15985584940@163.com;504387687@qq.com;liqinghe@caas.cn
CLC Number:
LIU Sha, SU Meng, GAO Qianmei, SONG Danli, ZHAO Guiping, LI Jianhui, LI Qinghe. Transcriptome Analysis of Chicken Macrophages after SIRT1 Activated[J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(6): 2661-2671.
Table 1
Primer sequences for RT-qPCR"
| 基因Gene | 上游引物(5′→3′)Forward | 下游引物(5′→3′)Reverse |
| β-actin | GAGAAATTGTGCGTGACATCA | CCTGAACCTCTCATTGCCA |
| CD14 | ACCCGACACTTGACCCTGTTG | TCTGCCTTAGGGGAAAAGGGC |
| NFKBIA | TGGGAGAACAGCACTACACT | GCCCTGGTAGGTCACTTTG |
| TLR7 | GCACACCGGAAAATGGTACA | TTGGGAAACCAACGTCCTGAT |
| JUN | AACTCCGCACCCAACTAC | GGTACAGTCTGAGGCTCTTCT |
| IL10RA | TGGGCATCTTCGACACTGAC | GAGTTGGTTTGCACCGTGAG |
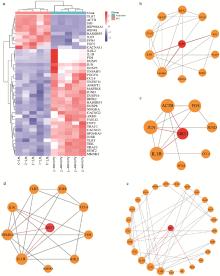
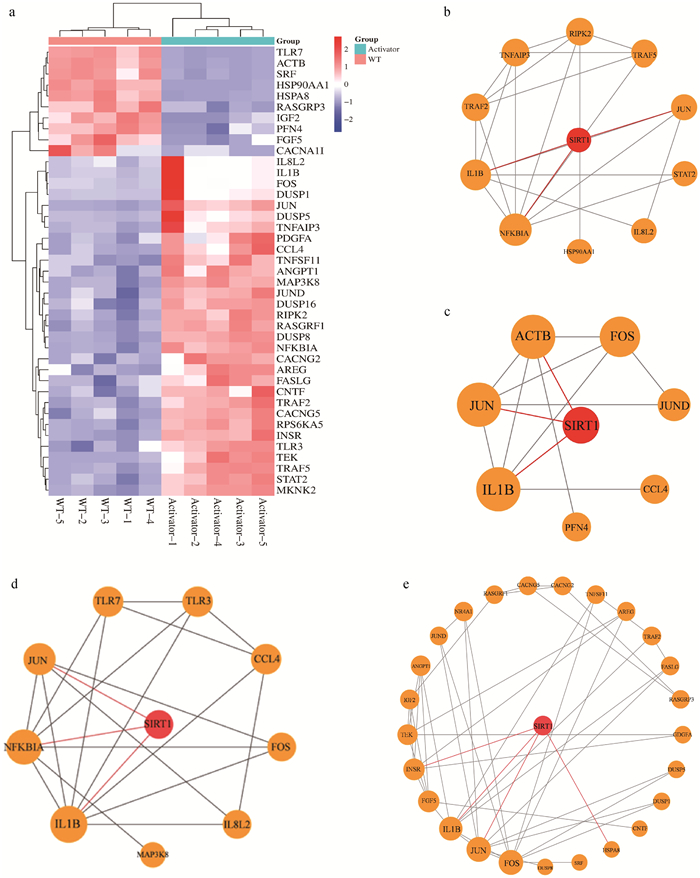
Fig. 3
Construction of proteins interaction network a. Hierarchical clustering diagram of differentially expressed genes in MAPK, NOD, Toll-like receptor signaling pathways, and Salmonella infection signaling pathways; b-e. SIRT1 and NOD, Salmonella, TOLL, MAPK signaling pathway differential genes interaction proteins network diagram"
| 1 |
YANG Y S , LIU Y , WANG Y W , et al. Regulation of SIRT1 and its roles in inflammation[J]. Front Immunol, 2022, 13, 831168.
doi: 10.3389/fimmu.2022.831168 |
| 2 |
TANG Z , WANG P , DONG C , et al. Oxidative stress signaling mediated pathogenesis of diabetic cardiomyopathy[J]. Oxid Med Cell Longev, 2022, 2022, 5913374.
doi: 10.1155/2022/5913374 |
| 3 |
XU C , WANG L , FOZOUNI P , et al. SIRT1 is downregulated by autophagy in senescence and ageing[J]. Nat Cell Biol, 2020, 22 (10): 1170- 1179.
doi: 10.1038/s41556-020-00579-5 |
| 4 |
SHARMA A , MAHUR P , MUTHUKUMARAN J , et al. Shedding light on structure, function and regulation of human sirtuins: a comprehensive review[J]. 3 Biotech, 2023, 13 (1): 29.
doi: 10.1007/s13205-022-03455-1 |
| 5 |
MIHANFAR A , AKBARZADEH M , GHAZIZADEH DARBAND S , et al. SIRT1: a promising therapeutic target in type 2 diabetes mellitus[J]. Arch Physiol Biochem, 2024, 130 (1): 13- 28.
doi: 10.1080/13813455.2021.1956976 |
| 6 |
LIU D , LIU X , MA X , et al. Two novel InDels within the promoter of SIRT1 are associated with growth traits in chickens[J]. Br Poult Sci, 2022, 63 (4): 445- 453.
doi: 10.1080/00071668.2021.2014400 |
| 7 |
CHANG N , LI J , LIN S , et al. Emerging roles of SIRT1 activator, SRT2104, in disease treatment[J]. Sci Rep, 2024, 14 (1): 5521.
doi: 10.1038/s41598-024-55923-8 |
| 8 |
REN J , XU N , MA Z , et al. Characteristics of expression and regulation of sirtuins in chicken (Gallus gallus)[J]. Genome, 2017, 60 (5): 431- 440.
doi: 10.1139/gen-2016-0125 |
| 9 |
SHEN Z , AJMO J M , ROGERS C Q , et al. Role of SIRT1 in regulation of LPS- or two ethanol metabolites-induced TNF-alpha production in cultured macrophage cell lines[J]. Am J Physiol Gastrointest Liver Physiol, 2009, 296 (5): G1047- 53.
doi: 10.1152/ajpgi.00016.2009 |
| 10 |
YEUNG F , HOBERG J E , RAMSEY C S , et al. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase[J]. EMBO J, 2004, 23 (12): 2369- 2380.
doi: 10.1038/sj.emboj.7600244 |
| 11 |
TENG Y , HUANG Y , YU H , et al. Nimbolide targeting SIRT1 mitigates intervertebral disc degeneration by reprogramming cholesterol metabolism and inhibiting inflammatory signaling[J]. Acta Pharm Sin B, 2023, 13 (5): 2269- 2280.
doi: 10.1016/j.apsb.2023.02.018 |
| 12 |
PARK S Y , LEE S W , LEE S Y , et al. SIRT1/Adenosine monophosphate-activated protein kinase α signaling enhances macrophage polarization to an anti-inflammatory phenotype in rheumatoid arthritis[J]. Front Immunol, 2017, 8, 1135.
doi: 10.3389/fimmu.2017.01135 |
| 13 | XU C Q , LI J , LIANG Z Q , et al. Sirtuins in macrophage immune metabolism: A novel target for cardiovascular disorders[J]. Int J Biol Macromol, 2024, 256 (Pt 1): 128270. |
| 14 |
ZHAO X , LI M , LU Y , et al. Sirt1 inhibits macrophage polarization and inflammation in gouty arthritis by inhibiting the MAPK/NF-κB/AP-1 pathway and activating the Nrf2/HO-1 pathway[J]. Inflamm Res, 2024, 73 (7): 1173- 1184.
doi: 10.1007/s00011-024-01890-9 |
| 15 |
DU N , WU K , ZHANG J , et al. Inonotsuoxide B regulates M1 to M2 macrophage polarization through sirtuin-1/endoplasmic reticulum stress axis[J]. Int Immunopharmacol, 2021, 96, 107603.
doi: 10.1016/j.intimp.2021.107603 |
| 16 |
MOURITS V P , HELDER L S , MATZARAKI V , et al. The role of sirtuin 1 on the induction of trained immunity[J]. Cell Immunol, 2021, 366, 104393.
doi: 10.1016/j.cellimm.2021.104393 |
| 17 |
ELESELA S , MORRIS S B , NARAYANAN S , et al. Sirtuin 1 regulates mitochondrial function and immune homeostasis in respiratory syncytial virus infected dendritic cells[J]. PLoS Pathog, 2020, 16 (2): e1008319.
doi: 10.1371/journal.ppat.1008319 |
| 18 |
LIN L , WEN S H , GUO S Z , et al. Role of SIRT1 in Streptococcus pneumoniae-induced human β-defensin-2 and interleukin-8 expression in A549 cell[J]. Mol Cell Biochem, 2014, 394 (1-2): 199- 208.
doi: 10.1007/s11010-014-2095-2 |
| 19 | 陈鑫鹏, 夏榕鸽, 盖新燕, 等. 冷刺激对APP感染仔猪肺损伤的影响及其作用机制研究[J]. 中国畜牧兽医, 2025, 52 (3): 1383- 1392. |
| CHEN X P , XIA R G , GAI X Y , et al. Effects and mechanism of cold stimulation on lung injury of APP-infected piglets.[J]. China Animal Husbandry & Veterinary Medicine, 2025, 52 (3): 1383- 1392. | |
| 20 |
GANESAN R , HOS N J , GUTIERREZ S , et al. Salmonella Typhimurium disrupts Sirt1/AMPK checkpoint control of mTOR to impair autophagy[J]. PLoS Pathog, 2017, 13 (2): e1006227.
doi: 10.1371/journal.ppat.1006227 |
| 21 |
LOVE M I , HUBER W , ANDERS S . Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2[J]. Genome Biol, 2014, 15 (12): 550.
doi: 10.1186/s13059-014-0550-8 |
| 22 | 陈洋. 牛卵泡发育相关基因筛选及SIRT1、INHBA基因功能研究[D]. 长春: 吉林农业大学2018. |
| CHEN Y. Selection of genes related to novine follicular development and studyon the functions of SIRT1 and INHBA gene. [D]. Changchun: Jilin Agricultural University, 2018. (in Chinese) | |
| 23 | 张琪, 张锦, 王杰, 等. 肠炎沙门菌感染鸡lncRNA表达谱分析[J]. 中国家禽, 2021, 43 (7): 18- 27. |
| ZHANG Q , ZHANG J , WANG J , et al. lncRNA expression profiles in chickens challenged by Salmonella Enteritidis[J]. China Poultry, 2021, 43 (7): 18- 27. | |
| 24 |
VINCENDEAU M , HADIAN K , MESSIAS A C , et al. Inhibition of canonical NF-κB signaling by a small molecule targeting NEMO-ubiquitin interaction[J]. Sci Rep, 2016, 6, 18934.
doi: 10.1038/srep18934 |
| 25 |
KUMAR M , SAHU S K , KUMAR R , et al. MicroRNA let-7 modulates the immune response to Mycobacterium tuberculosis infection via control of A20, an inhibitor of the NF-κB pathway[J]. Cell Host Microbe, 2015, 17 (3): 345- 356.
doi: 10.1016/j.chom.2015.01.007 |
| 26 |
KANG H , KIM S C , OH Y . Fucoxanthin Abrogates Ionizing Radiation-Induced Inflammatory Responses by Modulating Sirtuin 1 in Macrophages[J]. Mar Drugs, 2023, 21 (12): 635.
doi: 10.3390/md21120635 |
| 27 |
HE S , WANG Y , LIU J , et al. Activating SIRT1 deacetylates NF-κB p65 to alleviate liver inflammation and fibrosis via inhibiting NLRP3 pathway in macrophages[J]. Int J Med Sci, 2023, 20 (4): 505- 519.
doi: 10.7150/ijms.77955 |
| 28 | JIAO F , GONG Z . The beneficial roles of SIRT1 in neuroinflammation-related diseases[J]. Oxid Med Cell Longev, 2020, 2020, 6782872. |
| 29 |
CHEN H , DENG J , GAO H , et al. Involvement of the SIRT1-NLRP3 pathway in the inflammatory response[J]. Cell Commun Signal, 2023, 21 (1): 185.
doi: 10.1186/s12964-023-01177-2 |
| 30 |
SAXTON R A , TSUTSUMI N , SU L L , et al. Structure-based decoupling of the pro- and anti-inflammatory functions of interleukin-10[J]. Science, 2021, 371 (6535): eabc8433.
doi: 10.1126/science.abc8433 |
| 31 | 余盼. 奶牛p38 MAPK信号通路相关因子表达与乳房炎的相关性分析[D]. 南京: 南京农业大学, 2015. |
| YU P. Correlation analysis between the p38 MAPK signaling pathway associsated molecule and bovine mastitis. [D]. Nanjing: Nanjing Agricultural University, 2015. (in Chinese) | |
| 32 |
YOSHIZAKI T , SCHENK S , IMAMURA T , et al. SIRT1 inhibits inflammatory pathways in macrophages and modulates insulin sensitivity[J]. Am J Physiol Endocrinol Metab, 2010, 298 (3): E419- 428.
doi: 10.1152/ajpendo.00417.2009 |
| 33 |
PARK S Y , LEE S W , LEE S Y , et al. SIRT1/Adenosine monophosphate-activated protein kinase α signaling enhances macrophage polarization to an anti-inflammatory phenotype in rheumatoid arthritis[J]. Front Immunol, 2017, 8, 1135.
doi: 10.3389/fimmu.2017.01135 |
| 34 | HAJRA D , RAJMANI R S , CHAUDHARY A D , et al. Salmonella-induced SIRT1 and SIRT3 are crucial for maintaining the metabolic switch in bacteria and host for successful pathogenesis[J]. Elife, 2024, 13. |
| 35 |
MÓTYÁN J A , BAGOSSI P , BENKŐ S , et al. A molecular model of the full-length human NOD-like receptor family CARD domain containing 5 (NLRC5) protein[J]. BMC Bioinformatics, 2013, 14, 275.
doi: 10.1186/1471-2105-14-275 |
| 36 |
EREN E , BERBER M , ÖZÖREN N . NLRC3 protein inhibits inflammation by disrupting NALP3 inflammasome assembly via competition with the adaptor protein ASC for pro-caspase-1 binding[J]. J Biol Chem, 2017, 292 (30): 12691- 12701.
doi: 10.1074/jbc.M116.769695 |
| 37 |
ZHANG L , MO J , SWANSON K V , et al. NLRC3, a member of the NLR family of proteins, is a negative regulator of innate immune signaling induced by the DNA sensor STING[J]. Immunity, 2014, 40 (3): 329- 341.
doi: 10.1016/j.immuni.2014.01.010 |
| 38 |
STAPELS D A C , HILL P W S , WESTERMANN A J , et al. Salmonella persisters undermine host immune defenses during antibiotic treatment[J]. Science, 2018, 362 (6419): 1156- 1160.
doi: 10.1126/science.aat7148 |
| 39 |
PANAGI I , JENNINGS E , ZENG J , et al. Salmonella effector SteE converts the mammalian serine/threonine kinase GSK3 into a tyrosine kinase to direct macrophage polarization[J]. Cell Host Microbe, 2020, 27 (1): 41- 53.e6.
doi: 10.1016/j.chom.2019.11.002 |
| [1] | LIU Zilong, LI Qiao, WU Yi, WANG Huihui, LI Taotao, MA Youji. Transcriptomics Reveals the Effects of Chinese Herbal Feed Additives on Bile Acids Metabolism and Immune Function in Hu Sheep Liver Tissue [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(6): 3014-3026. |
| [2] | SU Meng, LIU Sha, SONG Danli, GAO Qianmei, ZHENG Maiqing, WEN Jie, ZHAO Guiping, LI Qinghe. Identification of Candidate Genes Associated with Ascites Syndrome in Broilers Based on Transcriptome Sequencing [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(2): 559-570. |
| [3] | ZHANG Zhengyu, YANG Peihong, GUO Hong, LI Xin, ZHANG Linlin, GUO Yiwen, HU Debao, DING Xiangbin. Effects of Sirt1 Deacetylase on Proliferation and Differentiation of Bovine Skeletal Muscle Satellite Cells [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(2): 603-610. |
| [4] | HU Hanwen, BAO Tugeqin, REN Xiujuan, DING Wenqi, GONG Wendian, JIA Zijie, SHI Lin, MA Muren, Baorigele , DUGARJAVIIN Manglai, BAI Dongyi. Comparative Study on Muscle Fiber Development Phenotype and Gene Expression Profile of Two Mongolian Horse Populations [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(2): 643-656. |
| [5] | WU Shuang, YIN Na, YU Mohan, PING Yuyu, BAI Hao, CHEN Shihao, CHANG Guobin. The Effect of TRIM39.2 Overexpression on the Transcriptional Expression of Chicken Macrophages [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(1): 178-188. |
| [6] | LU Xiu, ZHANG Ming'ai, KONG Min, ZHANG Jing, WANG Binghan, HOU Zhongyi, TENG Xingyi, JIANG Yajing, FAN Wenlei, WANG Baowei. Screening for Candidate Genes Related to Egg Production in Wulong Geese Based on Transcriptome and Proteome Analyses [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(1): 232-245. |
| [7] | Xiaoxu ZHANG, Hao LI, Pingjie FENG, Hao YANG, Xinyue LI, Ran LÜ, Zhangyuan PAN, Mingxing CHU. Application of Single-Cell Transcriptome Sequencing Technology in Domesticated Animals [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(8): 3276-3287. |
| [8] | Jing CHEN, Xuebei WU, Dongzhi MIAO, Chi ZHANG, Zhenyu GUO, Ying WANG. Comparative Analysis of Transcriptome of Pigeon Follicles at Early Stage of Laying Interval Reveals Genes Related to Follicular Development [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(8): 3503-3515. |
| [9] | Wanqing LI, Yaqi ZENG, Xinkui YAO, Jianwen WANG, Xinxin YUAN, Chen MENG, Yuanfang SUN, Xuan PENG, Jun MENG. Comparative Analysis of Blood Transcriptome in Yili Horses Bred for Meat Performance [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(7): 2951-2962. |
| [10] | Mingliang HE, Xiaoyang LÜ, Yongqing JIANG, Zhenghai SONG, Yeqing WANG, Huiguo YANG, Shanhe WANG, Wei SUN. Function Analysis of SOX18 in Hu Sheep Hair Follicle Dermal Papilla Cells Based on Transcriptome Sequencing [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(6): 2409-2420. |
| [11] | CHEN Zhe, QU Xiaolu, GUO Binbin, SUN Xuefeng, YAN Leyan. Study on Candidate Genes for Green Light Affecting Early Development of Goose Embryo Heart Based on Transcriptome Sequencing [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(5): 1978-1988. |
| [12] | XU Junjie, ZHANG Lutong, WANG Jinjie, CHEN Xiaochen, HE Weixian, CAI Chuanjiang, CHU Guiyan, YANG Gongshe. Exploring the Effect of Epimedium on Estrus of Gilts Based on Multiomics and Network Pharmacology [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(4): 1615-1628. |
| [13] | WANG Xin, NIE Tong, LI Aqun, MA Jun. Hesperidin Alleviates High-fat-diet Induced Hepatic Oxidative Stress in Mice via Oxidative Phosphorylation Pathway [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(3): 1302-1313. |
| [14] | GAO Yawei, PENG Di, SUN Zhaoyang, YAN Ziyue, CUI Kai, MA Zefang. Mining the Molecular Mechanism of Exogenous Melatonin Affecting the Development of Mink Ovary Based on Transcriptome Data [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(2): 607-618. |
| [15] | MIN Xiangyu, WEI Jiali, XU Biao, LIU Huitao, ZHENG Junjun, WANG Guiwu. Full-length Transcriptome Sequencing of Sika Deer Antler and Mining of Antler Yield-related Genes [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(12): 5549-5566. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||