





畜牧兽医学报 ›› 2025, Vol. 56 ›› Issue (3): 1147-1158.doi: 10.11843/j.issn.0366-6964.2025.03.016
王红1,2( ), 赵为民3, 程金花3, 李惠侠2,*(
), 赵为民3, 程金花3, 李惠侠2,*( ), 方晓敏1,*(
), 方晓敏1,*( )
)
收稿日期:2024-10-11
出版日期:2025-03-23
发布日期:2025-04-02
通讯作者:
李惠侠,方晓敏
E-mail:2749363442@qq.com;lihuixia@njau.edu.cn;fxmw2000@163.com
作者简介:王红(2000-),女,安徽宿州人,硕士生,主要从事猪分子遗传育种研究,E-mail: 2749363442@qq.com
基金资助:
WANG Hong1,2( ), ZHAO Weimin3, CHENG Jinhua3, LI Huixia2,*(
), ZHAO Weimin3, CHENG Jinhua3, LI Huixia2,*( ), FANG Xiaomin1,*(
), FANG Xiaomin1,*( )
)
Received:2024-10-11
Online:2025-03-23
Published:2025-04-02
Contact:
LI Huixia, FANG Xiaomin
E-mail:2749363442@qq.com;lihuixia@njau.edu.cn;fxmw2000@163.com
摘要:
旨在鉴定猪CYP3A29基因核心启动子及相应的转录调控因子,分析转录因子对CYP3A29启动子活性的调控。本研究以3头健康的大白母猪(30 kg)为试验材料,利用PCR和Western blot检测CYP3A29基因在猪各组织(心、肝、脾、肺、肾、小肠、肌肉)中的表达分布;构建不同片段长度的CYP3A29基因启动子双荧光素酶报告载体,转染293T和AML12细胞系,检测荧光素酶活性,确定CYP3A29基因的核心启动子区域;利用Animal TFDB网站分析CYP3A29核心启动子区域可能存在的转录调控因子,针对核心启动子区域构建分段缺失双荧光素酶报告载体,检测荧光素酶活性大小,确定转录因子结合位点;构建转录因子结合突变位点的双荧光素酶报告载体和转录因子shRNA载体,探讨转录因子对CYP3A29核心启动子的调控作用。结果显示,CYP3A29基因在猪肝脏中表达量最高;CYP3A29启动子4个不同检测区域(-2 026~+62 bp、-1 526~+62 bp、-1 026~+62 bp和-528~+62 bp)中-528~+62 bp活性最高,为CYP3A29核心启动子区;CYP3A29启动子-528~-448 bp区域负向调控核心启动子活性,且含有潜在的转录因子RUNX1结合位点;突变RUNX1结合位点可显著降低-528~+62 bp启动子的荧光素酶活性,而干扰RUNX1基因则显著升高-528~+62 bp野生型启动子的荧光素酶活性,但对-528~+62 bp突变型启动子的荧光素酶活性无显著影响,提示RUNX1转录因子可负向调控CYP3A29基因核心启动子活性。本研究结果为进一步解析猪CYP3A29基因的转录调控机制奠定了基础。
中图分类号:
王红, 赵为民, 程金花, 李惠侠, 方晓敏. 猪CYP3A29基因核心启动子鉴定及转录调控分析[J]. 畜牧兽医学报, 2025, 56(3): 1147-1158.
WANG Hong, ZHAO Weimin, CHENG Jinhua, LI Huixia, FANG Xiaomin. Identification and Transcriptional Regulation Analysis of the Core Promoter of Porcine CYP3A29 Gene[J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(3): 1147-1158.
表 1
CYP3A29启动子分段扩增引物信息"
| 引物Primer | 引物序列(5′→3′)Primer sequence | 片段长度/bp Length |
| PGL-CYP3A29(-2 026~+62)-F1 | GCTG$\underline{{\rm{GCTAGC}}}$ TCTACTTCATGGTGCCCAGAG | 2 088 |
| PGL-CYP3A29(-1 526~+62)-F2 | GCTG$\underline{{\rm{GCTAGC}}}$GAGCTGGGACTCACTGCATG | 1 588 |
| PGL-CYP3A29(-1 026~+62)-F3 | GCTG$\underline{{\rm{GCTAGC}}}$TTCCTGCGTCACACCCTTCA | 1 088 |
| PGL-CYP3A29(-528~+62)-F4 | GCTC$\underline{{\rm{GCTAGC}}}$AATTGTGCTCAGGCGGATAG | 590 |
| PGL-CYP3A29(-448~+62)-F5 | GCTC$\underline{{\rm{GCTAGC}}}$AAGATAGACTATTCTTTCTGAGC | 510 |
| PGL-CYP3A29(-289~+62)-F6 | GCTC$\underline{{\rm{GCTAGC}}}$GGAGACATGGCATTTTATAGG | 351 |
| PGL-CYP3A29(-96~+62)-F7 | GCTC$\underline{{\rm{GCTAGC}}}$TGGTGTTTTTTCACTGGCTGC | 158 |
| PGL-CYP3A29-promoter-R | CCCAAGCTTGCCACTGTCCTCGTGATTCT | |
| CYP3A29 | F: ACATCTTTGGGGCCTACAGC R: AGATCGGGGTGAGGAATGGA | 177 |
| HPRT(pig) | F: TGACCAGTCAACGGGCGATA R: CAACACTTCGAGGGGTCCTT | 197 |
| HPRT(mouse) | F: CCATCACATTGTGGCCCTCT R: TTATGTCCCCCGTTGACTGA | 167 |
| RUNX1 | F: TGGCAGGCAACGATGAAAAC R: GCAACTTGTGGCGGATTTGT | 163 |
| Spi1 | F: GAACCACTTCACAGAGCTGC R: GTCATCTTCTTGCGGTTGCC | 474 |
| Spib | F: CAGAGGACTTCACCAGCCAG R: GAGGAGAACTGGAAGACGCC | 232 |
表 2
RUNX1基因干扰载体引物信息"
| 引物Primer | 引物序列(5′→3′)Primer sequence |
| shRUNX1-F | CACC$\underline{{\rm{GCAGAACTGAGAAATGCTACC}}}$CTCGAG$\underline{{\rm{GGTAGCATTTCTCAGTTCTG}}}$CTTTTTT |
| shRUNX1-R | AAACAAAAAA$\underline{{\rm{GCAGAACTGAGAAATGCTACC}}}$CTCGAG$\underline{{\rm{GGTAGCATTTCTCAGTTCTGC}}}$ |
| shNC-F | CACCG$\underline{{\rm{TTCTCCGAACGTGTCACGT}}}$CTCGAG$\underline{{\rm{ACGTGACACGTTCGGAGAA}}}$CTTTTTT |
| shNC-R | AAACAAAAAAG$\underline{{\rm{TTCTCCGAACGTGTCACGT}}}$CTCGAG$\underline{{\rm{ACGTGACACGTTCGGAGAA}}}$C |
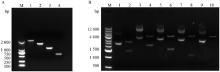
图 2
CYP3A29启动子分段扩增与载体双酶切验证 A.CYP3A29基因启动子分段扩增: 1~4.分别代表CYP3A29基因启动子-2 026~+62 bp、-1 526~+62 bp、-1 026~+62 bp和-528~ +62 bp的扩增片段电泳结果;B. CYP3A29基因启动子荧光素酶报告载体的双酶切结果验证: 1~2.pRL-TK质粒与双酶切验证;3~4.pGL4.10-CYP3A29(-2 026~+62 bp) 质粒与双酶切验证;5~6.pGL4.10-CYP3A29(-1 526~+62 bp) 质粒与双酶切验证;7~8.pGL4.10-CYP3A29(-1 026~+62 bp) 质粒与双酶切验证;9~10.pGL4.10-CYP3A29(-528~+62 bp) 质粒与双酶切验证"


图 4
CYP3A29基因核心启动子的分段活性分析 A. CYP3A29基因核心启动子的分段扩增:1~3.CYP3A29基因核心启动子-448~+62 bp、-289~+62 bp和-96~+62 bp的扩增片段。B. CYP3A29基因启动子荧光素酶报告载体的双酶切结果验证:1~2.pGL4.10-CYP3A29(-448~ +62 bp) 质粒与双酶切验证;3~4. pGL4.10-CYP3A29(-289~+62 bp) 质粒与双酶切验证;5~6.pGL4.10-CYP3A29(-96~+62 bp) 质粒与双酶切验证。C. CYP3A29基因核心分段启动子在AML12细胞的表达活性"

| 1 |
ZHANG L Y , XU X Q , BADAWY S , et al. A review: effects of macrolides on CYP450 enzymes[J]. Curr Drug Metab, 2020, 21 (12): 928- 937.
doi: 10.2174/1389200221666200817113920 |
| 2 |
LIN S Q , WEI J C , YANG B T . Bioremediation of organic pollutants by white rot fungal cytochrome P450: the role and mechanism of CYP450 in biodegradation[J]. Chemosphere, 2022, 301, 134776.
doi: 10.1016/j.chemosphere.2022.134776 |
| 3 |
LOOS N H C , BEIJNEN J H , SCHINKEL A H . The mechanism-based inactivation of CYP3A4 by ritonavir: what mechanism?[J]. Int J Mol Sci, 2022, 23 (17): 9866.
doi: 10.3390/ijms23179866 |
| 4 | LOLODI O , WANG Y M , WRIGHT W C , et al. Differential regulation of CYP3A4 and CYP3A5 and its implication in drug discovery[J]. Curr Drug Metab, 2017, 18 (12): 1095- 1105. |
| 5 |
ZANGER U M , SCHWAB M . Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation[J]. Pharmacol Ther, 2013, 138 (1): 103- 141.
doi: 10.1016/j.pharmthera.2012.12.007 |
| 6 |
WOODLAND C , HUANG T T , GRYZ E , et al. Expression, activity and regulation of CYP3A in human and rodent brain[J]. Drug Metab Rev, 2008, 40 (1): 149- 168.
doi: 10.1080/03602530701836712 |
| 7 |
ACHOUR B , BARBER J , ROSTAMI-HODJEGAN A . Cytochrome P450 Pig liver pie: determination of individual cytochrome P450 isoform contents in microsomes from two pig livers using liquid chromatography in conjunction with mass spectrometry[J]. Drug Metab Dispos, 2011, 39 (11): 2130- 2134.
doi: 10.1124/dmd.111.040618 |
| 8 |
HE Y C , ZHOU X Q , LI X W , et al. Relationship between CYP3A29 and pregnane X receptor in landrace pigs: pig CYP3A29 has a similar mechanism of regulation to human CYP3A4[J]. Comp Biochem Physiol Part C Toxicol Pharmacol, 2018, 214, 9- 16.
doi: 10.1016/j.cbpc.2018.08.006 |
| 9 |
YAO M , DAI M H , LIU Z Y , et al. Comparison of the substrate kinetics of pig CYP3A29 with pig liver microsomes and human CYP3A4[J]. Biosci Rep, 2011, 31 (3): 211- 220.
doi: 10.1042/BSR20100084 |
| 10 |
SOUCEK P , ZUBER R , ANZENBACHEROVÁ E , et al. Minipig cytochrome P450 3A, 2A and 2C enzymes have similar properties to human analogs[J]. BMC Pharmacol, 2001, 1 (1): 11.
doi: 10.1186/1471-2210-1-11 |
| 11 |
ANZENBACHEROVÁ E , BARANOVÁ J , ZUBER R , et al. Model systems based on experimental animals for studies on drug metabolism in man: (mini)pig cytochromes P450 3A29 and 2E1[J]. Basic Clin Pharmacol Toxicol, 2005, 96 (3): 244- 245.
doi: 10.1111/j.1742-7843.2005.pto960316.x |
| 12 |
LI X W , JIN X Q , ZHOU X L , et al. Pregnane X receptor is required for IFN-α-mediated CYP3A29 expression in pigs[J]. Biochem Biophys Res Commun, 2014, 445 (2): 469- 474.
doi: 10.1016/j.bbrc.2014.02.011 |
| 13 |
LI X W , HU X Z , JIN X E , et al. IFN-γ regulates cytochrome 3A29 through pregnane X receptor in pigs[J]. Xenobiotica, 2015, 45 (5): 373- 379.
doi: 10.3109/00498254.2014.985761 |
| 14 |
PASZTI-GERE E , MATIS G , FARKAS O , et al. The effects of intestinal LPS exposure on inflammatory responses in a porcine enterohepatic co-culture system[J]. Inflammation, 2014, 37 (1): 247- 260.
doi: 10.1007/s10753-013-9735-7 |
| 15 | YANG X Y , XING F , WANG L , et al. Effect of pregnane X receptor on CYP3A29 expression in porcine alveolar macrophages during Mycoplasma hyopneumoniae infection[J]. Animals (Basel), 2021, 11 (2): 349. |
| 16 |
方晓敏, 赵为民, 付言峰, 等. 猪支原体肺炎发生的品种敏感差异及分子基础[J]. 中国农业科学, 2015, 48 (14): 2839- 2847.
doi: 10.3864/j.issn.0578-1752.2015.14.015 |
|
FANG X M , ZHAO W M , FU Y F , et al. Difference in susceptibility to mycoplasma pneumonia among various pig breeds and its molecular genetic basis[J]. Scientia Agricultura Sinica, 2015, 48 (14): 2839- 2847.
doi: 10.3864/j.issn.0578-1752.2015.14.015 |
|
| 17 | JOVER R , BORT R , GÓMEZ-LECHÓN M J , et al. Down-regulation of human CYP3A4 by the inflammatory signal interleukin 6: molecular mechanism and transcription factors involved[J]. FASEB J, 2002, 16 (13): 1799- 1801. |
| 18 |
LE CARPENTIER E C , CANET E , MASSON D , et al. Impact of inflammation on midazolam metabolism in severe COVID-19 patients[J]. Clin Pharmacol Ther, 2022, 112 (5): 1033- 1039.
doi: 10.1002/cpt.2698 |
| 19 |
KASARLA S S , GARIKAPATI V , KUMAR Y , et al. Interplay of vitamin D and CYP3A4 polymorphisms in endocrine disorders and cancer[J]. Endocrinol Metab (Seoul), 2022, 37 (3): 392- 407.
doi: 10.3803/EnM.2021.1349 |
| 20 |
HUA R , QIAO G J , CHEN G S , et al. Single-cell RNA-sequencing analysis of colonic lamina propria immune cells reveals the key immune cell-related genes of ulcerative colitis[J]. J Inflamm Res, 2023, 16, 5171- 5188.
doi: 10.2147/JIR.S440076 |
| 21 |
SHEN W K , CHEN S Y , GAN Z Q , et al. AnimalTFDB 4.0: a comprehensive animal transcription factor database updated with variation and expression annotations[J]. Nucleic Acids Res, 2023, 51 (D1): D39- D45.
doi: 10.1093/nar/gkac907 |
| 22 |
PAN Z Y , YAO Y L , YIN H W , et al. Pig genome functional annotation enhances the biological interpretation of complex traits and human disease[J]. Nat Commun, 2021, 12 (1): 5848.
doi: 10.1038/s41467-021-26153-7 |
| 23 |
ZHAO Y X , HOU Y , XU Y Y , et al. A compendium and comparative epigenomics analysis of cis-regulatory elements in the pig genome[J]. Nat Commun, 2021, 12 (1): 2217.
doi: 10.1038/s41467-021-22448-x |
| 24 |
LI J J , XIANG Y , ZHANG L , et al. Enhancer-promoter interaction maps provide insights into skeletal muscle-related traits in pig genome[J]. BMC Biol, 2022, 20 (1): 136.
doi: 10.1186/s12915-022-01322-2 |
| 25 | QIN W P , PAN J P , QIN Y W , et al. Identification of functional glucocorticoid response elements in the mouse FoxO1 promoter[J]. Biochem Biophys Res Commun, 2014, 450 (2): 979- 983. |
| 26 | ZHANG Y , ZHU F , TENG J , et al. Effects of salinity stress on methylation of the liver genome and complement gene in large yellow croaker (Larimichthys crocea)[J]. Fish Shellfish Immunol, 2022, 129, 207- 220. |
| 27 | ZHU M J , LV J H , WANG W , et al. CMPK2 is a host restriction factor that inhibits infection of multiple coronaviruses in a cell-intrinsic manner[J]. PLoS Biol, 2023, 21 (3): e3002039. |
| 28 | WEI F X , GU Y , HE L Z , et al. HSD17B6 delays type 2 diabetes development via inhibiting SREBP activation[J]. Metabolism, 2023, 145, 155631. |
| 29 | ZHU H F , ZHANG R Z , YI L , et al. UNC93B1 attenuates the cGAS-STING signaling pathway by targeting STING for autophagy-lysosome degradation[J]. J Med Virol, 2022, 94 (9): 4490- 4501. |
| 30 | YANG M L , CAI W S , LIN Z H , et al. Intermittent hypoxia promotes TAM-induced glycolysis in laryngeal cancer cells via regulation of HK1 expression through activation of ZBTB10[J]. Int J Mol Sci, 2023, 24 (19): 14808. |
| 31 | CHENG M Y , KANYEMA M M , SUN Y , et al. African swine fever virus L83L negatively regulates the cGAS-STING-mediated IFN-Ⅰ pathway by recruiting tollip to promote STING autophagic degradation[J]. J Virol, 2023, 97 (2): e01923-22. |
| 32 | GUO X , ZHANG Z Y , LIN C H , et al. A/(H1N1) pdm09 NS1 promotes viral replication by enhancing autophagy through hijacking the IAV negative regulatory factor LRPPRC[J]. Autophagy, 2023, 19 (5): 1533- 1550. |
| 33 | WU Y K , CHEN W J , MIAO H L , et al. SIRT7 promotes the proliferation and migration of anaplastic thyroid cancer cells by regulating the desuccinylation of KIF23[J]. BMC Cancer, 2024, 24 (1): 210. |
| 34 | ZHANG G H , LI D Y , TU C F , et al. Loss-of-function missense variant of AKAP4 induced male infertility through reduced interaction with QRICH2 during sperm flagella development[J]. Hum Mol Genet, 2021, 31 (2): 219- 231. |
| 35 | CHEN Y Z , HE Y Y , LIU S B . RUNX1-regulated signaling pathways in ovarian cancer[J]. Biomedicines, 2023, 11 (9): 2357. |
| 36 | TANG X J , ZHONG L C , TIAN X , et al. RUNX1 promotes mitophagy and alleviates pulmonary inflammation during acute lung injury[J]. Sig Transduct Target Ther, 2023, 8 (1): 288. |
| 37 | ARIFFIN N S . RUNX1 as a novel molecular target for breast cancer[J]. Clin Breast Cancer, 2022, 22 (6): 499- 506. |
| 38 | HONG D L , FRITZ A J , GORDON J A , et al. RUNX1-dependent mechanisms in biological control and dysregulation in cancer[J]. J Cell Physiol, 2019, 234 (6): 8597- 8609. |
| 39 | REED-INDERBITZIN E , MORENO-MIRALLES I , VANDEN-EYNDEN S K , et al. RUNX1 associates with histone deacetylases and SUV39H1 to repress transcription[J]. Oncogene, 2006, 25 (42): 5777- 5786. |
| 40 | TELFER J C , HEDBLOM E E , ANDERSON M K , et al. Localization of the domains in runx transcription factors required for the repression of CD4 in thymocytes[J]. J Immunol, 2004, 172 (7): 4359- 4370. |
| [1] | 刘爱军, 张传亮, 黄晓兵, 周彩琴. 猪繁殖与呼吸综合征病毒生命周期的研究进展[J]. 畜牧兽医学报, 2025, 56(3): 1027-1041. |
| [2] | 邬沛伶, 李依璇, 王浩杰, 李亚菲, 刘绍蒙, 刘青芸, 王湘如. 猪流行性腹泻疫苗研究进展[J]. 畜牧兽医学报, 2025, 56(3): 1042-1058. |
| [3] | 黄雅妮, 唐熹, 李井泉, 魏嘉诚, 吴珍芳, 李新云, 肖石军, 张志燕. 大规模群体解析猪日增重及达百千克体重日龄的潜在因果基因[J]. 畜牧兽医学报, 2025, 56(3): 1100-1109. |
| [4] | 吴嘉浩, 吴姿仪, 窦腾飞, 白利瑶, 张永前, 董联合, 李鹏飞, 李新建, 韩雪蕾, 李秀领. 豫农黑猪生长相关性状的拷贝数变异全基因组关联分析研究[J]. 畜牧兽医学报, 2025, 56(3): 1110-1119. |
| [5] | 杨宇婷, 陈国梁, 常巧宁, 鲍武, 刘靖超, 姬梦婷, 荣晓音, 郭晓红, 杨阳, 李步高. miR-375-3p靶向Fam229a调控猪前体脂肪细胞分化[J]. 畜牧兽医学报, 2025, 56(3): 1120-1133. |
| [6] | 贾万里, 王继英, 李菁璇, 王彦平, 耿立英, 张传生, 赵雪艳. 基于转录组测序技术鉴别影响莱芜猪滴水损失的关键基因[J]. 畜牧兽医学报, 2025, 56(3): 1134-1146. |
| [7] | 周泰增, 杨祎挺, 朱悦华, 钱洪喜, 刘一辉, 甘麦邻, 朱砺, 沈林園. 母猪死胎和木乃伊全基因组关联分析[J]. 畜牧兽医学报, 2025, 56(3): 1231-1241. |
| [8] | 刘晨龙, 季华员, 卢丹, 万明春, 胡耀, 周泉勇. FST对猪卵巢颗粒细胞增殖凋亡及激素分泌的影响[J]. 畜牧兽医学报, 2025, 56(3): 1242-1251. |
| [9] | 周文涛, 王晨昱, 周辉, 刘洪彪, 冯舒锾, 范高升, 李铁军, 何流琴. 单宁酸对免疫应激断奶仔猪肌肉形态、风味氨基酸及肌纤维相关基因表达的影响[J]. 畜牧兽医学报, 2025, 56(3): 1290-1301. |
| [10] | 张越, 茹毅, 郝荣增, 杨锐, 赵陇和, 李亚军, 杨洋, 张荣, 蒋成辉, 郑海学. 非洲猪瘟病毒H108R蛋白的制备及其免疫原性评价[J]. 畜牧兽医学报, 2025, 56(3): 1344-1354. |
| [11] | 马晓莉, 李段, 曾道平, 刘燕玲, 王晓敏, 彭国良, 宋长绪, 王磊, 徐铮. 非洲猪瘟病毒p72蛋白抗体全自动化学发光酶免疫检测方法的建立[J]. 畜牧兽医学报, 2025, 56(3): 1355-1365. |
| [12] | 余昕雅, 何海健, 王磊, 倪语晨, 杜静, 周莹珊, 董婉玉, 王晓杜. LncRNA 18850对猪流行性腹泻病毒复制的影响[J]. 畜牧兽医学报, 2025, 56(3): 1366-1375. |
| [13] | 范杰, 邰易润, 朱艳丽, 陈志雄, 胡巧云, 陈智, 刘甜甜, 李欣, 范仲鑫, 葛猛. 近年湖南省猪圆环病毒2型感染状况的调查及遗传演化分析[J]. 畜牧兽医学报, 2025, 56(3): 1376-1385. |
| [14] | 潘俊毅, 吴青瑶, 谭碧娥, 郭秋平, 黄瑞林, 陈家顺. 生长育肥猪精准营养供给技术及智能化养殖设备研究进展[J]. 畜牧兽医学报, 2025, 56(2): 501-512. |
| [15] | 张冬萱, 王智豪, 乔岩, 赵肖肖, 范松杰, 张超. 猪流行性腹泻病毒S1蛋白的原核表达及其核酸适配体的筛选[J]. 畜牧兽医学报, 2025, 56(2): 839-850. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
