





Acta Veterinaria et Zootechnica Sinica ›› 2025, Vol. 56 ›› Issue (4): 1608-1620.doi: 10.11843/j.issn.0366-6964.2025.04.012
• Review • Previous Articles Next Articles
LI Xiaohan1,2( ), LI Guiping2(
), LI Guiping2( ), HUO Caiyun2, ZHANG Qilong3, SUN Yingjian1, SUN Huiling2,*(
), HUO Caiyun2, ZHANG Qilong3, SUN Yingjian1, SUN Huiling2,*( )
)
Received:2024-05-27
Online:2025-04-23
Published:2025-04-28
Contact:
SUN Huiling
E-mail:1025290934@qq.com;lgp709@sina.cn;sunhuiling01@163.com
CLC Number:
LI Xiaohan, LI Guiping, HUO Caiyun, ZHANG Qilong, SUN Yingjian, SUN Huiling. Class II CRISPR/Cas Systems and Their Applications in Bacterial Synthetic Biology[J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(4): 1608-1620.
Table 1
The classification of class II CRISPR systems"
| 类型 Type | Ⅱ型 Type Ⅱ | Ⅴ型 Type Ⅴ | Ⅵ型 Type Ⅵ |
| 标志性效应蛋白 Signature effector protein | Cas9,II-A | Cas12a,V-A | Cas13a,VI-A |
| 发源菌 Bacteria of origin | 化脓性链球菌 | 新凶手弗朗西斯菌氨基酸球菌 | 纤毛菌瘤胃球菌 |
| 氨基酸球菌 | 瘤胃球菌 | ||
| 毛螺菌 | 普雷沃菌 | ||
| 摩拉菌 | 李斯特菌 | ||
| CRISPR-RNA | sgRNA,100 nt | crRNA,42~44 nt | crRNA,52~66 nt |
| PAM序列 | G富集 | T富集 | 无 |
| PAM sequence | 5′-NGG-3′ | 5′-TT(T)N-3′ | PFS序列 |
| 核酸酶结构域Nuclease domain | HNH、RuvC | RuvC | HEPN |
| 切割方式Cutting mode | dsDNA,平末端 | dsDNA,黏末端 | ssRNA |
| RNA酶活性RNAse activity | 无 | 有 | 有 |
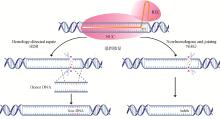
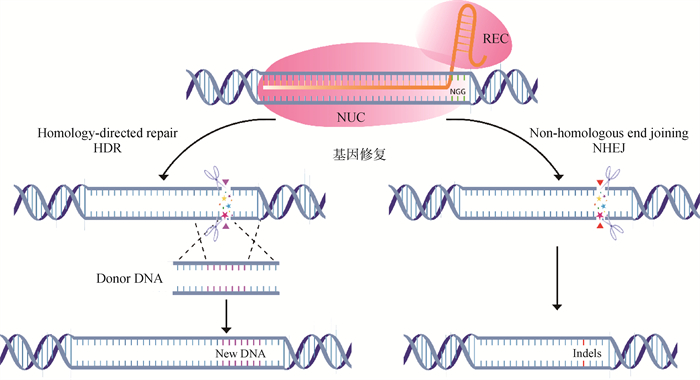
Fig. 2
The gene repair pattern diagram of the class Ⅱ CRISPR system NUC. Nucleolytic cleavage domain; REC. crRNA recognition domain; NGG. PAM recognition region; HDR. Homology-directed repair; NHEJ. Non-homologous end joining; Donor DNA. Homologous repair template; New DNA. Newly synthesized strand; Indels. Insertion-deletion mutations"
| 1 |
CHANDRASEGARAN S , CARROLL D . Origins of programmable nucleases for genome engineering[J]. J Mol Biol, 2016, 428 (5): 963- 989.
doi: 10.1016/j.jmb.2015.10.014 |
| 2 |
DEVEAU H , GARNEAU J E , MOINEAU S . CRISPR/Cas system and its role in phage-bacteria interactions[J]. Annu Rev Microbiol, 2010, 64, 475- 493.
doi: 10.1146/annurev.micro.112408.134123 |
| 3 |
ISHINO Y , SHINAGAWA H , MAKINO K , et al. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product[J]. J Bacteriol, 1987, 169 (12): 5429- 5433.
doi: 10.1128/jb.169.12.5429-5433.1987 |
| 4 |
JANSEN R , VAN EMBDEN J D A , GAASTRA W , et al. Identification of genes that are associated with DNA repeats in prokaryotes[J]. Mol Microbiol, 2002, 43 (6): 1565- 1575.
doi: 10.1046/j.1365-2958.2002.02839.x |
| 5 |
JINEK M , CHYLINSKI K , FONFARA I , et al. A programmable dual RNA-guided DNA endonuclease in adaptive bacterial immunity[J]. Science, 2012, 337 (6096): 816- 821.
doi: 10.1126/science.1225829 |
| 6 |
MAKAROVA K S , HAFT D H , BARRANGOU R , et al. Evolution and classification of the CRISPR-Cas systems[J]. Nat Rev Microbiol, 2011, 9 (6): 467- 477.
doi: 10.1038/nrmicro2577 |
| 7 |
MAKAROVA K S , WOLF Y I , KOONIN E V . The basic building blocks and evolution of CRISPR-Cas systems[J]. Biochem Soc Trans, 2013, 41 (6): 1392- 1400.
doi: 10.1042/BST20130038 |
| 8 |
MAKAROVA K S , WOLF Y I , ALKHNBASHI O S , et al. An updated evolutionary classification of CRISPR-Cas systems[J]. Nat Rev Microbiol, 2015, 13 (11): 722- 736.
doi: 10.1038/nrmicro3569 |
| 9 |
ZHAO J , FANG H , ZHANG D W . Expanding application of CRISPR-Cas9 system in microorganisms[J]. Synth Syst Biotechnol, 2020, 5 (4): 269- 276.
doi: 10.1016/j.synbio.2020.08.001 |
| 10 |
TAKEDA S N , NAKAGAWA R , OKAZAKI S , et al. Structure of the miniature type V-F CRISPR-Cas effector enzyme[J]. Mol Cell, 2021, 81 (3): 558- 570.
doi: 10.1016/j.molcel.2020.11.035 |
| 11 |
GAO P , YANG H , RAJASHANKAR K R , et al. Type V CRISPR-Cas Cpf1 endonuclease employs a unique mechanism for crRNA-mediated target DNA recognition[J]. Cell Res, 2016, 26 (8): 901- 913.
doi: 10.1038/cr.2016.88 |
| 12 |
VAN BELJOUW S P B , SANDERS J , RODRÍGUEZ-MOLINA A , et al. RNA-targeting CRISPR-Cas systems[J]. Nat Rev Microbiol, 2023, 21 (1): 21- 34.
doi: 10.1038/s41579-022-00793-y |
| 13 |
SHMAKOV S , ABUDAYYEH O O , MAKAROVA K S , et al. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems[J]. Mol Cell, 2015, 60 (3): 385- 397.
doi: 10.1016/j.molcel.2015.10.008 |
| 14 |
GARNEAU J E , DUPUIS M È , VILLION M , et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA[J]. Nature, 2010, 468 (7320): 67- 71.
doi: 10.1038/nature09523 |
| 15 |
CHYLINSKI K , MAKAROVA K S , CHARPENTIER E , et al. Classification and evolution of type II CRISPR-Cas systems[J]. Nucleic Acids Res, 2014, 42 (10): 6091- 6105.
doi: 10.1093/nar/gku241 |
| 16 |
LIEBER M R . The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway[J]. Annu Rev Biochem, 2010, 79, 181- 211.
doi: 10.1146/annurev.biochem.052308.093131 |
| 17 |
ZHANG J P , LI X L , LI G H , et al. Efficient precise knockin with a double cut HDR donor after CRISPR/Cas9-mediated double-stranded DNA cleavage[J]. Genome Biol, 2017, 18 (1): 35.
doi: 10.1186/s13059-017-1164-8 |
| 18 |
HIDALGO-CANTABRANA C , GOH Y J , BARRANGOU R . Characterization and repurposing of type I and type II CRISPR-Cas systems in bacteria[J]. J Mol Biol, 2019, 431 (1): 21- 33.
doi: 10.1016/j.jmb.2018.09.013 |
| 19 |
HELER R , SAMAI P , MODELL J W , et al. Cas9 specifies functional viral targets during CRISPR-Cas adaptation[J]. Nature, 2015, 519 (7542): 199- 202.
doi: 10.1038/nature14245 |
| 20 |
LARSON M H , GILBERT L A , WANG X W , et al. CRISPR interference (CRISPRi) for sequence-specific control of gene expression[J]. Nat Protoc, 2013, 8 (11): 2180- 2196.
doi: 10.1038/nprot.2013.132 |
| 21 |
CLETO S , JENSEN J V K , WENDISCH V F , et al. Corynebacterium glutamicum metabolic engineering with CRISPR interference (CRISPRi)[J]. ACS Synth Biol, 2016, 5 (5): 375- 385.
doi: 10.1021/acssynbio.5b00216 |
| 22 | SONG X , HUANG H , XIONG Z Q , et al. CRISPR-Cas9D10A nickase-assisted genome editing in Lactobacillus casei[J]. Appl Environ Microbiol, 2017, 83 (22): e01259- 17. |
| 23 |
LIU Z Q , DONG H N , CUI Y L , et al. Application of different types of CRISPR/Cas-based systems in bacteria[J]. Microb Cell Fact, 2020, 19 (1): 172.
doi: 10.1186/s12934-020-01431-z |
| 24 |
LIU Y , WAN X Y , WANG B J . Engineered CRISPRa enables programmable eukaryote-like gene activation in bacteria[J]. Nat Commun, 2019, 10 (1): 3693.
doi: 10.1038/s41467-019-11479-0 |
| 25 | GOH Y J , BARRANGOU R . Portable CRISPR-Cas9N system for flexible genome engineering in Lactobacillus acidophilus, Lactobacillus gasseri, and Lactobacillus paracasei[J]. Appl Environ Microbiol, 2021, 87 (6): e02669- 20. |
| 26 |
CHEN Q L , WANG B , PAN L . Efficient expression of γ-glutamyl transpeptidase in Bacillus subtilis via CRISPR/Cas9n and its immobilization[J]. Appl Microbiol Biotechnol, 2024, 108 (1): 149.
doi: 10.1007/s00253-023-12889-3 |
| 27 |
ZETSCHE B , GOOTENBERG J S , ABUDAYYEH O O , et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system[J]. Cell, 2015, 163 (3): 759- 771.
doi: 10.1016/j.cell.2015.09.038 |
| 28 |
ZHONG Z H , ZHANG Y X , YOU Q , et al. Plant genome editing using FnCpf1 and LbCpf1 Nucleases at redefined and altered PAM sites[J]. Mol Plant, 2018, 11 (7): 999- 1002.
doi: 10.1016/j.molp.2018.03.008 |
| 29 |
SAFARI F , ZARE K , NEGAHDARIPOUR M , et al. CRISPR Cpf1 proteins: structure, function and implications for genome editing[J]. Cell Biosci, 2019, 9, 36.
doi: 10.1186/s13578-019-0298-7 |
| 30 |
ZHU X W , WU Y K , LV K Q , et al. Combining CRISPR-Cpf1 and recombineering facilitates fast and efficient genome editing in Escherichia coli[J]. ACS Synth Biol, 2022, 11 (5): 1897- 1907.
doi: 10.1021/acssynbio.2c00041 |
| 31 |
ZHANG Y , YUAN J F . CRISPR/Cas12a-mediated genome engineering in the photosynthetic bacterium Rhodobacter capsulatus[J]. Microb Biotechnol, 2021, 14 (6): 2700- 2710.
doi: 10.1111/1751-7915.13805 |
| 32 |
KNOOT C J , BISWAS S , PAKRASI H B . Tunable repression of key photosynthetic processes using Cas12a CRISPR interference in the fast-growing cyanobacterium Synechococcus sp. UTEX 2973[J]. ACS Synth Biol, 2020, 9 (1): 132- 143.
doi: 10.1021/acssynbio.9b00417 |
| 33 |
YOU Y , ZHANG P P , WU G S , et al. Highly specific and sensitive detection of Yersinia pestis by portable Cas12a-UPTLFA platform[J]. Front Microbiol, 2021, 12, 700016.
doi: 10.3389/fmicb.2021.700016 |
| 34 |
SAM I K , CHEN Y Y , MA J , et al. TB-QUICK: CRISPR-Cas12b-assisted rapid and sensitive detection of Mycobacterium tuberculosis[J]. J Infect, 2021, 83 (1): 54- 60.
doi: 10.1016/j.jinf.2021.04.032 |
| 35 |
ABUDAYYEH O O , GOOTENBERG J S , KONERMANN S , et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector[J]. Science, 2016, 353 (6299): aaf5573.
doi: 10.1126/science.aaf5573 |
| 36 |
EAST-SELETSKY A , O'CONNELL M R , KNIGHT S C , et al. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection[J]. Nature, 2016, 538 (7624): 270- 273.
doi: 10.1038/nature19802 |
| 37 |
GARNER K L . Principles of synthetic biology[J]. Essays Biochem, 2021, 65 (5): 791- 811.
doi: 10.1042/EBC20200059 |
| 38 | 蒋成辉, 曾巧英, 王萌, 等. CRISPR/Cas9构建srtA基因敲除的金黄色葡萄球菌[J]. 生物技术通报, 2020, 36 (9): 253- 265. |
| JIANG C H , ZENG Q Y , WANG M , et al. Construction of srtA-knockout strain in Staphylococcus aureus by CRISPR/Cas9 technology[J]. Biotechnology Bulletin, 2020, 36 (9): 253- 265. | |
| 39 | 杨茗崴, 王玮玉, 王裕光, 等. fimH基因敲除对大肠杆菌NMGCF-19菌株致病性的影响[J]. 中国兽医学报, 2023, 43 (9): 1881- 1887. |
| YANG M W , WANG W Y , WANG Y G , et al. Effect of fimH gene knockout on the pathogenicity of Escherichia coli NMGCF-19 strain[J]. Chinese Journal of Veterinary Science, 2023, 43 (9): 1881- 1887. | |
| 40 | 何京桦, 乐科易. 利用CRISPR/Cas9系统敲除大肠杆菌tnaA基因[J]. 生物学杂志, 2019, 36 (6): 17- 19. |
| HE J H , LE K Y . CRISPR/ Cas9-mediated tnaA knockout in Escherichia coli[J]. Journal of Biology, 2019, 36 (6): 17- 19. | |
| 41 | RAINHA J , RODRIGUES J L , RODRIGUES L R . CRISPR-Cas9:a powerful tool to efficiently engineer Saccharomyces cerevisiae[J]. Life (Basel), 2020, 11 (1): 13. |
| 42 |
GÖTTL V L , SCHMITT I , BRAUN K , et al. CRISPRi-library-guided target identification for engineering carotenoid production by Corynebacterium glutamicum[J]. Microorganisms, 2021, 9 (4): 670.
doi: 10.3390/microorganisms9040670 |
| 43 |
XIN Q L , CHEN Y D , CHEN Q L , et al. Development and application of a fast and efficient CRISPR-based genetic toolkit in Bacillus amyloliquefaciens LB1ba02[J]. Microb Cell Fact, 2022, 21 (1): 99.
doi: 10.1186/s12934-022-01832-2 |
| 44 |
AMERUOSO A , VILLEGAS KCAM M C , COHEN K P , et al. Activating natural product synthesis using CRISPR interference and activation systems in Streptomyces[J]. Nucleic Acids Res, 2022, 50 (13): 7751- 7760.
doi: 10.1093/nar/gkac556 |
| 45 | 张博, 杨玉凤, 贺周霖, 等. 利用CRISPRi挖掘大肠杆菌生物合成D-泛酸的关键基因[J]. 微生物学报, 2023, 63 (12): 4625- 4643. |
| ZHANG B , YANG Y F , HE Z L , et al. Mining key genes for biosynthesis of D-pantothenic acid in Escherichia coli by CRISPRi[J]. Acta Microbiologica Sinica, 2023, 63 (12): 4625- 4643. | |
| 46 | HAO M J , HE Y Z , ZHANG H F , et al. CRISPR-Cas9-mediated carbapenemase gene and plasmid curing in carbapenem-resistant Enterobacteriaceae[J]. Antimicrob Agents Chemother, 2020, 64 (9): e00843- 20. |
| 47 |
WU X , ZHA J , KOFFAS M A G , et al. Reducing Staphylococcus aureus resistance to lysostaphin using CRISPR-dCas9[J]. Biotechnol Bioeng, 2019, 116 (12): 3149- 3159.
doi: 10.1002/bit.27143 |
| 48 |
WAN X L , LI Q Y , OLSEN R H , et al. Engineering a CRISPR interference system targeting AcrAB-TolC efflux pump to prevent multidrug resistance development in Escherichia coli[J]. J Antimicrob Chemother, 2022, 77 (8): 2158- 2166.
doi: 10.1093/jac/dkac166 |
| 49 | CHEN S Z , CAO H L , XU Z R , et al. A type I-F CRISPRi system unveils the novel role of CzcR in modulating multidrug resistance of Pseudomonas aeruginosa[J]. Microbiol Spectr, 2023, 11 (5): e01123- 23. |
| 50 |
GU T N , ZHAO S Q , PI Y S , et al. Highly efficient base editing in Staphylococcus aureus using an engineered CRISPR RNA-guided cytidine deaminase[J]. Chem Sci, 2018, 9 (12): 3248- 3253.
doi: 10.1039/C8SC00637G |
| 51 | 吕言, 任晓昕, 徐大庆. CRISPR/Cas9和λ-Red基因敲除技术在大肠杆菌中的应用比较[J]. 微生物前沿, 2022, 11 (1): 1- 10. |
| LÜ Y , REN X X , XU D Q . Comparative study of CRISPR/Cas9 and λ-red gene knockout techniques in Eschrichia coli[J]. Advances in Microbiology, 2022, 11 (1): 1- 10. | |
| 52 |
JIANG Y , CHEN B , DUAN C L , et al. Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system[J]. Appl Environ Microbiol, 2015, 81 (7): 2506- 2514.
doi: 10.1128/AEM.04023-14 |
| 53 |
SU B L , SONG D D , ZHU H H . Homology-dependent recombination of large synthetic pathways into E. coli genome via λ-Red and CRISPR/Cas9 dependent selection methodology[J]. Microb Cell Fact, 2020, 19 (1): 108.
doi: 10.1186/s12934-020-01360-x |
| 54 |
ZHAO R , LIU Y Q , ZHANG H , et al. CRISPR-Cas12a-mediated gene deletion and regulation in Clostridium ljungdahlii and its application in carbon flux redirection in synthesis gas fermentation[J]. ACS Synth Biol, 2019, 8 (10): 2270- 2279.
doi: 10.1021/acssynbio.9b00033 |
| 55 |
KO Y J , CHA J , JEONG W Y , et al. Bio-isopropanol production in Corynebacterium glutamicum: metabolic redesign of synthetic bypasses and two-stage fermentation with gas stripping[J]. Bioresour Technol, 2022, 354, 127171.
doi: 10.1016/j.biortech.2022.127171 |
| 56 | YAN M Y , YAN H Q , REN G X , et al. CRISPR-Cas12a-assisted recombineering in bacteria[J]. Appl Environ Microbiol, 2017, 83 (17): e00947- 17. |
| 57 |
AO X , YAO Y , LI T , et al. A multiplex genome editing method for Escherichia coli based on CRISPR-Cas12a[J]. Front Microbiol, 2018, 9, 2307.
doi: 10.3389/fmicb.2018.02307 |
| 58 | 黄紫蕾, 龙腾飞, 刘雅红, 等. 基于CRISPR-Cas12f对抗耐药革兰氏阴性菌的新型递送系统[C]//中国畜牧兽医学会兽医药理毒理学分会第十三次全国会员代表大会暨第十七次学术研讨会论文集. 南宁: 中国畜牧兽医学会兽医药理毒理学分会, 2023: 1. |
| HUANG Z L, LONG T F, LIU Y H, et al. A novel delivery system based on CRISPR-Cas12f for combating antibiotic-resistant Gram-negative bacteria[C]//Proceedings of the Chinese Association of Animal Science and Veterinary Medicine. Nanning: Chinese Society of Animal Husbandry and Veterinary Medicine Veterinary Pharmacology and Toxicology Branch, 2023: 1. (in Chinese) | |
| 59 |
AI J W , ZHOU X , XU T , et al. CRISPR-based rapid and ultra-sensitive diagnostic test for Mycobacterium tuberculosis[J]. Emerg Microbes Infect, 2019, 8 (1): 1361- 1369.
doi: 10.1080/22221751.2019.1664939 |
| 60 |
LI Y , DENG F , HALL T , et al. CRISPR/Cas12a-powered immunosensor suitable for ultra-sensitive whole Cryptosporidium oocyst detection from water samples using a plate reader[J]. Water Res, 2021, 203, 117553.
doi: 10.1016/j.watres.2021.117553 |
| 61 |
冀霞, 代绍密, 余若菁, 等. CRISPR-Cas12a检测牛奶中大肠杆菌方法的建立与评价[J]. 食品研究与开发, 2023, 44 (18): 179- 184.
doi: 10.12161/j.issn.1005-6521.2023.18.024 |
|
JI X , DAI S M , YU R J , et al. Establishment and evaluation of CRISPR-Cas12a method for detecting Escherichia coli in milk[J]. Food Research and Development, 2023, 44 (18): 179- 184.
doi: 10.12161/j.issn.1005-6521.2023.18.024 |
|
| 62 |
HAO J , XIE L F , YANG T M , et al. Naked-eye on-site detection platform for Pasteurella multocida based on the CRISPR-Cas12a system coupled with recombinase polymerase amplification[J]. Talanta, 2023, 255, 124220.
doi: 10.1016/j.talanta.2022.124220 |
| 63 |
LU L Y , ZHANG H M , LIN F H , et al. Sensitive and automated detection of bacteria by CRISPR/Cas12a-assisted amplification with digital microfluidics[J]. Sens Actuators B: Chem, 2023, 381, 133409.
doi: 10.1016/j.snb.2023.133409 |
| 64 |
COX D B T , GOOTENBERG J S , ABUDAYYEH O O , et al. RNA editing with CRISPR-Cas13[J]. Science, 2017, 358 (6366): 1019- 1027.
doi: 10.1126/science.aaq0180 |
| 65 |
KIGA K , TAN X E , IBARRA-CHÁVEZ R , et al. Development of CRISPR-Cas13a-based antimicrobials capable of sequence-specific killing of target bacteria[J]. Nat Commun, 2020, 11 (1): 2934.
doi: 10.1038/s41467-020-16731-6 |
| 66 |
GOOTENBERG J S , ABUDAYYEH O O , LEE J W , et al. Nucleic acid detection with CRISPR-Cas13a/C2c2[J]. Science, 2017, 356 (6336): 438- 442.
doi: 10.1126/science.aam9321 |
| 67 |
ZHOU J , YIN L J , DONG Y N , et al. CRISPR-Cas13a based bacterial detection platform: sensing pathogen Staphylococcus aureus in food samples[J]. Anal Chim Acta, 2020, 1127, 225- 233.
doi: 10.1016/j.aca.2020.06.041 |
| 68 |
LIU J , CARMICHAEL C , HASTURK H , et al. Rapid specific detection of oral bacteria using Cas13-based SHERLOCK[J]. J Oral Microbiol, 2023, 15 (1): 2207336.
doi: 10.1080/20002297.2023.2207336 |
| 69 | 侯雅超, 邢微微, 王亚楠, 等. 针对副溶血性弧菌toxR基因的RPA-CRISPR/Cas13a荧光快速检测方法的建立及应用[J]. 中华医院感染学杂志, 2024, 34 (14): 2081- 2086. |
| HOU Y C , XING W W , WANG Y N , et al. Establishment and application of RPA-CRISPR/Cas13a fluorescent rapid assay targeting Vibrio parahaemolyticus toxR gene[J]. Chinese Journal of Nosocomiology, 2024, 34 (14): 2081- 2086. |
| [1] | QIU Qian, SANG Rui, WANG Wei, LIU Xinman, YU Minghong, LIU Xiaotong, YU Tian, ZHANG Xuemei. Study on the Activity of Huning Powder against Chicken Lung-derived E. coli and the in vitro Effects of Anti-inflammation and Anti-oxidation [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(4): 1969-1980. |
| [2] | ZHAO Wenyue, YANG Jing, SHAO Yilan, LI Jiaxuan, JIANG Yanping, CUI Wen, WANG Xiaona, TANG Lijie. Screening and Identification of Secretory Signal Peptide of Lactobacillus reuteri Expressing Lactoferrin Peptide [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(3): 1431-1440. |
| [3] | BAO Binwu, ZOU Huiying, LI Junliang, GAO Chen, GAO Huijiang, DU Zhenwei, ZHANG Boyu, LI Junya, GAO Xue. Research Progress in Gene Editing Technology [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(1): 1-14. |
| [4] | ZHANG Lei, CHEN Liang, FENG Wanyu, LAN Shijie, MIAO Yan, TIAN Qiufeng, BAI Changsheng, ZHANG Bei, DONG Jiaqiang, JIANG Botao, WANG Hongbao, SHI Tongrui, HUANG Xuankai. The Role of Biofilms in the Pathogenesis of Animal Bacterial Infections [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(1): 107-114. |
| [5] | Wenwen LIU, Faming DONG, Yanzhen BI. The Development of Multi-Gene Editing Technology and Its Application in Agricultural Biological Germplasm Innovation [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(8): 3267-3275. |
| [6] | Ruiying LIANG, Jingxia SUO, Lin LIANG, Xianyong LIU, Jiabo DING, Xun SUO, Xinming TANG. Genetic Manipulation of Eimeria: Platform Development, Application, and Perspective [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(8): 3362-3373. |
| [7] | Bibo LI, Ke WU, Xiaolong SHI, Yining YAN, Jiahao LI, Guoqing DUAN, Xiong LI, Yanpeng REN, Jianing DONG, Chunxiang ZHANG, Youshe REN. Sheep-derived Lactobacillus plantarum Regulates the Bacterial Community and Mucosal Barrier in Jejunum of Diarrheic Lambs [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(8): 3552-3569. |
| [8] | Wenjun XU, Nan ZHENG, Jiaqi WANG, Lu MENG. Study on the Community Structure of Psychrophilic Bacteria in Raw Milk from North Regions of China based on Metagenomic Sequencing [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(8): 3659-3668. |
| [9] | Sijia LIU, Nan ZHENG, Jiaqi WANG, Shengguo ZHAO. Effects of Red Clover Extract on Microbial Diversity and Urea Decomposition in Rumen Fermentation of Dairy Cows in vitro [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(6): 2510-2518. |
| [10] | Huijie REN, Xun MA, Jing WANG, Caixia LIU, Dongdong ZENG, Lijun KOU, Weidi SHI, Shuangfei LÜ, Ruixuan QIAN, Shengjie GAO. Construction and Partial Biological Characteristics Trial of Lm4b_02325/26 Double Gene Deletion Strain of Listeria monocytogenes [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(6): 2578-2587. |
| [11] | Ming LI, Hongwei CUI, Jie GAO, Lele AN, Songli LI, Zhenghua RAO. Identification and Genomic Analysis of Pathogenic Escherichia coli in Small Intestinal Content of White Feather Broilers [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(6): 2692-2700. |
| [12] | HUANG Jie, RUAN Zihao, CAI Rui. Advances of the Application of Antimicrobial Peptides in the Preservation of Porcine Semen at Room Temperature [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(4): 1401-1411. |
| [13] | GAO Yuanji, LIU Chang, CHEN Miao, CHEN Songbiao, ZHANG Junfeng, LI Jing, JIA Yanyan, LIAO Chengshui, GUO Rongxian, DING Ke, YU Zuhua, SHANG Ke. Structure, Secretory Characteristics, and Pathogenic Mechanism of Bacterial Outer Membrane Vesicles [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(3): 971-983. |
| [14] | WANG Kang, LIU Geyan, WANG Yu, YANG Zhen, TANG Xinwei, CAO Sanjie, HUANG Xiaobo, YAN Qigui, WU Rui, ZHAO Qin, DU Senyan, WEN Xintian, WEN Yiping. Preparation of Glaesserella parasuis Ghosts Vaccine Delivering Porcine Circovirus Type 2 DNA Vaccine and Evaluation of Its Immunoprotective Effect in Mice [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(3): 1179-1191. |
| [15] | FU Xuezhen, LI Xincan, QIAN Hongyu, LÜ Hong, WU Chanyu, WANG Xiaohan, WANG Xiaohua, WANG Zhiying, ZHOU Zuoyong. Antibacterial Effect and Mechanism of Bergamot Essential Oil on Corynebacterium pseudotuberculosis [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(3): 1217-1227. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||