





Acta Veterinaria et Zootechnica Sinica ›› 2025, Vol. 56 ›› Issue (11): 5545-5562.doi: 10.11843/j.issn.0366-6964.2025.11.016
• Animal Biotechnology and Reproduction • Previous Articles Next Articles
MA Yuan1( ), JIN Haoyan1, WANG Nana1, XIE Yaru1, LI Tianjiao2, ZHANG Lingkai1,*(
), JIN Haoyan1, WANG Nana1, XIE Yaru1, LI Tianjiao2, ZHANG Lingkai1,*( )
)
Received:2025-03-25
Online:2025-11-23
Published:2025-11-27
Contact:
ZHANG Lingkai
E-mail:myuan1809@163.com;zhanglklab2023@163.com
CLC Number:
MA Yuan, JIN Haoyan, WANG Nana, XIE Yaru, LI Tianjiao, ZHANG Lingkai. Dynamic Transcriptional Differences in Porcine Round Spermatids Resolved Based on scRNA-seq[J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(11): 5545-5562.
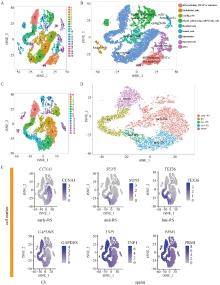
Fig. 2
Classification and cell type identification of porcine testes A. Porcine testis single-cell transcriptome data t-SNE and clustering analysis of cells divided into clusters; B. t-SNE and clustering analysis of cell types for each taxon of porcine testicular cells; C. Porcine testis spermatozoa t-SNE and recolonization analysis of cell division clusters; D. t-SNE and clustering analysis of cell types for each taxon of porcine testicular spermatids; E. Visualization of selected marker genes expression in all single cells in the t-SNE plot.1 or 2 cellular markers per cell population are showed"

| 1 |
DE R D . The nature and dynamics of spermatogonial stem cells[J]. Development, 2017, 144 (17): 3022- 3030.
doi: 10.1242/dev.146571 |
| 2 | HERMO L , PELLETIER R M , CYR D G , et al. Surfing the wave, cycle, life history, and genes/proteins expressed by testicular germ cells. Part 1: background to spermatogenesis, spermatogonia, and spermatocytes[J]. Microsc Res Tech, 2009, 73 (4): 241- 278. |
| 3 | 卜治文, 陆璐洋, 白顺, 等. 精子发生转录调控机制的研究进展[J]. 中国细胞生物学学报, 2024, 46 (4): 541- 552. |
| BU Z W , LU L Y , BAI S , et al. Advances in Transcriptional Regulation of Spermatogenesis[J]. Chinese Journal of Cell Biology, 2024, 46 (4): 541- 552. | |
| 4 | 张文杰, 陈琪, 周好乐, 等. 圆形及长形精子发生的相关基因[J]. 国际遗传学杂志, 2015, 38 (3): 166- 173. |
| ZHANG W J , CHEN Q , ZHOU H L , et al. Spermatogenesis related genes: circular and elongated spermatids[J]. International Journal of Genetics, 2015, 38 (3): 166- 173. | |
| 5 | 毛献宝. 人睾丸单倍体圆形精子细胞鉴别技术研究进展[J]. 中国临床新医学, 2020, 13 (11): 1178- 1182. |
| MAO X B . Research progress in identification techniques of human testis haploid round spermatid[J]. China Journal of New Clinical Medicine, 2020, 13 (11): 1178- 1182. | |
| 6 |
LI Z , LIU X , ZHANG Y , et al. FBXO24 modulates mRNA alternative splicing and MIWI degradation and is required for normal sperm formation and male fertility[J]. Elife, 2024, 12, RP91666.
doi: 10.7554/eLife.91666.3 |
| 7 |
PALERMO G , JORIS H , DEVROEY P , et al. Pregnancies after intracytoplasmic injection of single spermatozoon into an oocyte[J]. Lancet, 1992, 340 (8810): 17- 18.
doi: 10.1016/0140-6736(92)92425-F |
| 8 |
TESARIK J , MENDOZA C . Spermatid injection into human oocytes. Ⅰ. Laboratory techniques and special features of zygote development[J]. Hum Reprod, 1996, 11 (4): 772- 779.
doi: 10.1093/oxfordjournals.humrep.a019253 |
| 9 |
YANG W , ZHAO F , CHEN M , et al. Identification and characterization of male reproduction-related genes in pig (Sus scrofa) using transcriptome analysis[J]. BMC Genomics, 2020, 21 (1): 381.
doi: 10.1186/s12864-020-06790-w |
| 10 |
SAITOU M , HAYASHI K . Mammalian in vitro gametogenesis[J]. Science, 2021, 374 (6563): eaaz6830.
doi: 10.1126/science.aaz6830 |
| 11 |
OKWUN O , IGBOELI G , FORD J , et al. Number and function of sertoli cells, number and yield of spermatogonia, and daily sperm production in three breeds of boar[J]. J Reprod Fertil, 1996, 107 (1): 137- 149.
doi: 10.1530/jrf.0.1070137 |
| 12 |
SAHARE M , SUYATNO , IMAI H . Recent advances of in vitro culture systems for spermatogonial stem cells in mammals[J]. Reprod Med Biol, 2018, 17 (2): 134- 142.
doi: 10.1002/rmb2.12087 |
| 13 |
刘源壹, 李昕俞, 巴音那木拉, 等. 单细胞转录组测序技术及其在动物繁殖中的应用进展[J]. 畜牧兽医学报, 2023, 54 (2): 421- 433.
doi: 10.11843/j.issn.0366-6964.2023.02.001 |
|
LIU Y Y , LI X Y , BAYINNAMULA , et al. Single-cell transcriptome sequencing technology and its application in animal reproduction[J]. Acta Veterinaria et Zootechnica Sinica, 2023, 54 (2): 421- 433.
doi: 10.11843/j.issn.0366-6964.2023.02.001 |
|
| 14 |
ZHANG L , LI F , LEI P , et al. Single-cell RNA-sequencing reveals the dynamic process and novel markers in porcine spermatogenesis[J]. J Anim Sci Biotechnol, 2021, 12 (1): 122.
doi: 10.1186/s40104-021-00638-3 |
| 15 |
ZHANG P , CHEN X , ZHENG Y , et al. Long-term propagation of porcine undifferentiated spermatogonia[J]. Stem Cells Dev, 2017, 26 (15): 1121- 1131.
doi: 10.1089/scd.2017.0018 |
| 16 |
STUART T , BUTLER A , HOFFMAN P , et al. Comprehensive integration of single-cell data[J]. Cell, 2019, 177 (7): 1888- 1902.
doi: 10.1016/j.cell.2019.05.031 |
| 17 |
LIN T , CHEN T , LIU J , et al. Extending the Mann-Whitney-Wilcoxon rank sum test to survey data for comparing mean ranks[J]. Stat Med, 2021, 40 (7): 1705- 1717.
doi: 10.1002/sim.8865 |
| 18 |
QIU X , MAO Q , TANG Y , et al. Reversed graph embedding resolves complex single-cell trajectories[J]. Nat Methods, 2017, 14 (10): 979- 982.
doi: 10.1038/nmeth.4402 |
| 19 | ITO K , MURPHY D . Application of ggplot2 to pharmacometric graphics[J]. CPT Pharmacometrics Syst Pharmacol, 2013, 2 (10): e79. |
| 20 |
GUO J , GROW EJ , MLCOCHOVA H , et al. The adult human testis transcriptional cell atlas[J]. Cell Res, 2018, 28 (12): 1141- 1157.
doi: 10.1038/s41422-018-0099-2 |
| 21 |
NIAN H , FAN C , LIAO S , et al. RNF151, a testis-specific RING finger protein, interacts with dysbindin[J]. Arch Biochem Biophys, 2007, 465 (1): 157- 163.
doi: 10.1016/j.abb.2007.05.013 |
| 22 |
HEN L , SONG J , ZHANG J , et al. Spermatogenic cell-specific SPACA4 is essential for efficient sperm-zona pellucida binding in vitro[J]. Front Cell Dev Biol, 2023, 11, 1204017.
doi: 10.3389/fcell.2023.1204017 |
| 23 |
FUJIHARA Y , HERBERG S , BLAHA A , et al. The conserved fertility factor SPACA4/Bouncer has divergent modes of action in vertebrate fertilization[J]. Proc Natl Acad Sci U S A, 2021, 118 (39): e2108777118.
doi: 10.1073/pnas.2108777118 |
| 24 |
ZHENG M , CHEN X , CUI Y , et al. TULP2, a new RNA-binding protein, is required for mouse spermatid differentiation and male fertility[J]. Front Cell Dev Biol, 2021, 9, 623738.
doi: 10.3389/fcell.2021.623738 |
| 25 |
ADHAM I , NAYERNIA K , BURKHARDT-GÖTTGES E , et al. Teratozoospermia in mice lacking the transition protein 2 (Tnp2)[J]. Mol Hum Reprod, 2001, 7 (6): 513- 520.
doi: 10.1093/molehr/7.6.513 |
| 26 |
ZHANG L K , MA H D , GUO M , et al. Dynamic transcriptional atlas of male germ cells during porcine puberty[J]. Zool Res, 2022, 43 (4): 600- 603.
doi: 10.24272/j.issn.2095-8137.2022.037 |
| 27 |
ZHAO Y D , YANG C X , DU Z Q . Integrated single cell transcriptome sequencing analysis reveals species-specific genes and molecular pathways for pig spermiogenesis[J]. Reprod Domest Anim, 2023, 58 (12): 1745- 1755.
doi: 10.1111/rda.14493 |
| 28 | STEGER K , WILHELM J , KONRAD L , et al. Both protamine-1 to protamine-2 mRNA ratio and Bcl2 mRNA content in testicular spermatids and ejaculated spermatozoa discriminate between fertile and infertile men[J]. Hum Reprod, 2008, 23 (1): 11- 16. |
| 29 | GRASSETTI D , PAOLI D , GALLO M , et al. Protamine-1 and -2 polymorphisms and gene expression in male infertility: an Italian study[J]. J Endocrinol Invest, 2012, 35 (10): 882- 888. |
| 30 |
PAN D , FENG D , DING H , et al. Effects of bisphenol A exposure on DNA integrity and protamination of mouse spermatozoa[J]. Andrology, 2020, 8 (2): 486- 496.
doi: 10.1111/andr.12694 |
| 31 |
HU R , JIANG X , LIU G , et al. Polygenic co-expression changes the testis growth, hormone secretion and spermatogenesis to prompt puberty in Hu sheep[J]. Theriogenology, 2022, 194, 116- 125.
doi: 10.1016/j.theriogenology.2022.09.025 |
| 32 | 李美宁, 马庆, 弓韬, 等. 小鼠Spata3蛋白结构和功能的生信分析及初鉴[J]. 中国实验动物学报, 2022, 30 (6): 767- 776. |
| LI M N , MA Q , GONG T , et al. Bioinformatics analysis and primary identification of the structure and function of mouse Spata3 protein[J]. Acta Laboratorium Animalis Scientia Sinica, 2022, 30 (6): 767- 776. | |
| 33 |
黄向月, 熊显荣, 张磊, 等. SPATA3基因克隆及其在牦牛和犏牛睾丸中的差异表达研究[J]. 畜牧兽医学报, 2020, 51 (2): 279- 287.
doi: 10.11843/j.issn.0366-6964.2020.02.009 |
|
HUANG X Y , XIONG X R , ZHANG L , et al. Cloning of SPATA3 gene and its differential exptrssion analysis in yak and cattle-yak testis[J]. Acta Veterinaria et Zootechnica Sinica, 2020, 51 (2): 279- 287.
doi: 10.11843/j.issn.0366-6964.2020.02.009 |
|
| 34 |
ZHANG Y , LIU G , HUANG L , et al. SUN5 interacts with nuclear membrane LaminB1 and cytoskeletal GTPase Septin12 mediating the sperm head-and-tail junction[J]. Mol Hum Reprod, 2024, 30 (7): gaae022.
doi: 10.1093/molehr/gaae022 |
| 35 |
LOW-KAM C , RHAINDS D , LO K , et al. Variants at the APOE/C1/C2/C4 locus modulate cholesterol efflux capacity independently of high-density lipoprotein cholesterol[J]. J Am Heart Assoc, 2018, 7 (16): e009545.
doi: 10.1161/JAHA.118.009545 |
| 36 |
JONG M , HOFKER M , HAVEKES L . Role of ApoCs in lipoprotein metabolism: functional differences between ApoC1, ApoC2, and ApoC3[J]. Arterioscler Thromb Vasc Biol, 1999, 19 (3): 472- 484.
doi: 10.1161/01.ATV.19.3.472 |
| 37 |
FRANÇA L , AVELAR G , ALMEIDA F . Spermatogenesis and sperm transit through the epididymis in mammals with emphasis on pigs[J]. Theriogenology, 2005, 63 (2): 300- 318.
doi: 10.1016/j.theriogenology.2004.09.014 |
| 38 |
CHEN X , CHE D , ZHANG P , et al. Profiling of miRNAs in porcine germ cells during spermatogenesis[J]. Reproduction, 2017, 154 (6): 789- 798.
doi: 10.1530/REP-17-0441 |
| 39 | 雷佩佩. 基于单细胞测序解析猪精原细胞的异质性[D]. 杨凌: 西北农林科技大学, 2021. |
| LEI P P. The analysis of heterogeneity in porcine spermatogonia based on the single cell sequencing[D]. Yangling: Northwest A&F University, 2021. (in Chinese) | |
| 40 | 熊鑫. 猪胚胎早期卵裂时间与其发育潜能的关系[D]. 南宁: 广西大学, 2019. |
| XIONG X. Relationship between early cleavage time and developmental potential of pig embryo[D]. Nanning: Guangxi University, 2019. (in Chinese) | |
| 41 |
GREEN C , MA Q , MANSKE G , et al. A comprehensive roadmap of murine spermatogenesis defined by single-cell RNA-Seq[J]. Dev Cell, 2018, 46 (5): 651- 667.
doi: 10.1016/j.devcel.2018.07.025 |
| 42 |
YANG H , MA J , WAN Z , et al. Characterization of sheep spermatogenesis through single-cell RNA sequencing[J]. FASEB J, 2021, 35 (2): e21187.
doi: 10.1096/fj.202001035RRR |
| 43 |
SHETTY J , WOLKOWICZ M , DIGILIO L , et al. SAMP14, a novel, acrosomal membrane-associated, glycosylphosphatidylinositol-anchored member of the Ly-6/urokinase-type plasminogen activator receptor superfamily with a role in sperm-egg interaction[J]. J Biol Chem, 2003, 278 (33): 30506- 30515.
doi: 10.1074/jbc.M301713200 |
| 44 |
HE X , ZHANG Y , MAO Z , et al. SUN5, a testis-specific nuclear membrane protein, participates in recruitment and export of nuclear mRNA in spermatogenesis[J]. ActaFUJI Biochim Biophys Sin (Shanghai), 2024, 56 (11): 1673- 1686.
doi: 10.3724/abbs.2024134 |
| 45 |
GIRAULT M S , DUPUIS S , IALY-RADIO C , et al. Deletion of the Spata3 gene induces sperm alterations and in vitro hypofertility in mice[J]. Int J Mol Sci, 2021, 22 (4): 1959.
doi: 10.3390/ijms22041959 |
| 46 | 赵婷婷. 睾丸特异性基因Spata21基因在小鼠中mRNA的表达[J]. 人人健康, 2016 (23): 27. |
| ZHAO T T . Expression of mRNA for the testis-specific gene Spata21 gene in mice[J]. Health For Everyone, 2016 (23): 27. | |
| 47 |
MALLA A B , BHANDARI R . IP6K1 is essential for chromatoid body formation and temporal regulation of Tnp2 and Prm2 expression in mouse spermatids[J]. J Cell Sci, 2017, 130 (17): 2854- 2866.
doi: 10.1242/jcs.204966 |
| 48 | 陈晓旭. tsRNAs在猪精子发生成熟过程中动态变化及其在胚胎发育中的功能研究[D]. 杨凌: 西北农林科技大学, 2020. |
| CHEN X X. The research on tsRNA dynamics during porcine spermatogenesis, maturation and tsRNA functions in embryonic development[D]. Yangling: Northwest A&F University, 2020. (in Chinese) | |
| 49 |
WANG T , GAO H , LI W , et al. Essential role of histone replacement and modifications in male fertility[J]. Front Genet, 2019, 10, 962.
doi: 10.3389/fgene.2019.00962 |
| 50 |
XIONG C , WEN Z , LI G . Histone Variant H3 variant in health and in disease[J]. Sci China Life Sci, 2016, 59 (3): 245- 256.
doi: 10.1007/s11427-016-5006-9 |
| 51 |
GROSS K , HANSON B , HOTALING J . Round spermatid injection[J]. Urol Clin North Am, 2020, 47 (2): 175- 183.
doi: 10.1016/j.ucl.2019.12.004 |
| [1] | QIU Hualongchuan, JIN Qianqian, XU Xiaohan, ZHOU Jing, CAI Chengzhi, LI Long. Establishment of Ten Swine Pathogens Detection Method based on Nanopore Sequencing [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(9): 4546-4558. |
| [2] | LIU Junjun, GUO Donghui, LIU Huanhuan, SONG Runze, ZHAO Saiya, YANG Junyao, WEI Zhanyong, XIANG Yuqiang, CHEN Liying. Rapid Visual Detection for PDCoV/TGEV IgG Antibodies Using Smartphone-Assisted Colorimetric Sensing Platform based on Immunomagnetic Beads [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(9): 4559-4571. |
| [3] | LI Huimin, LEI Mingkai, RUAN Shengnan, LI Panpan, LI Wentao, HE Qigai. Establishment of Fluorescent Microsphere Immunochromatographic Assay for Porcine Epidemic Diarrhea Virus Antigen Detection [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(9): 4572-4580. |
| [4] | GUO Guihai, MA Rumeng, YIN Fangjie, LIU Xinzi, WANG Zi, MENG Weijing, LI Jiaxuan, CUI Wen, JIANG Yanping, TANG Lijie, ZHAO Haiyuan, WANG Xiaona. Study on the Expression of Recombinant Limosilactobacillus reuteri Expressing the S1 Gene of Porcine Epidemic Diarrhea Virus to Induce Specific Immune Responses in Piglets [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(9): 4581-4592. |
| [5] | TAO Lihan, LIN Cui, WU Chengcheng, KANG Zhaofeng, HUANG Jianzhen. Research Progress on the Structure and Function of Proteins Encoded by Porcine Deltacoronavirus [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(8): 3678-3689. |
| [6] | HU Jinling, ZHONG Qiqi, HUANG Cheng, LEI Minggang. AKR1B1 Regulates Proliferation and Differentiation of Porcine Skeletal Muscle Satellite Cells via the AMPK/mTOR/S6 Signaling Pathway [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(8): 3722-3733. |
| [7] | LI Kang, CHEN Siying, SUN Yawen, LENG Xuan, WANG Dong, CUI Kai, PANG Yunwei. Effects of Betaine on Preimplantation Development of Porcine Parthenogenetic Embryos [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(8): 3826-3836. |
| [8] | CHEN Bingbing, CAI Weiyou, LIU Yutong, WANG Xiuwu, SUN Shouhu, HE Dongsheng. Establishment and Application of Droplet Digital PCR Method for the Detection of Porcine Rotavirus A [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(8): 4095-4100. |
| [9] | TIAN Ru, FU Xingwei, HU Leyu, ZHU Mingjun, TONG Dewen. Isolation and Pathogenicity Analysis of a GⅡa Porcine Epidemic Diarrhea Virus [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(8): 4101-4111. |
| [10] | WANG Feiyan, LIU Chaofan, ZHANG Ya'nan, ZHOU Xiaotian, REN Jing, YUAN Chen, LI Tanqing, SONG Qinye. Preparation of Monoclonal Antibody to Porcine IL-15 and Establishment of Double Antibody Sandwich ELISA Method [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(7): 3442-3452. |
| [11] | LI Zhiqiang, CHEN Xueqing, ZHANG Yuanshu. Detection of Angiotensin Converting Enzyme 2 in Intestinal Tissues of Clinically Infected Porcine Epidemic Diarrhea Virus Piglets and Analysis of Its Relationship with Intestinal Pathological Changes [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(7): 3463-3473. |
| [12] | RUI Xue, ZHANG Yunhang, LI Yang, TAN Chen, CAI Yifei, LIU Yuanyuan, CAO Zongxi, ZHANG Yan, SUN Ruiping, LIU Guangliang. Establishment of a Triple PCR Method for the Detection of Porcine Endogenous Retroviruses and Its Preliminary Application to the Detection of Wuzhishan Pig Tissue Samples [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(7): 3548-3554. |
| [13] | WANG Yunke, WANG Na, YUE Ke, HE Kunmiao, ZHANG Xing, LIU Yao, ZHANG Gaiping. Substances with Inhibitory Effects on Porcine Epidemic Diarrhea Virus Replication in vitro [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(6): 2577-2589. |
| [14] | WU Chao, MING Wenhan, LU Shuwan, YANG Caimei, LIU Jinsong, MA Xiang, ZHANG Ruiqiang. Innate Immune Evasion Mechanisms of Porcine Epidemic Diarrhea Virus and Advances in Prevention and Control Strategies [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(6): 2590-2599. |
| [15] | ZHOU Min, TANG Deyuan, ZENG Zhiyong, WANG Bin, HUANG Tao, HU Wenwen, MAO Yinming, ZHOU Piao, HE Song. Research Progress on the Interaction between Porcine Epidemic Diarrhea Virus Proteins and Host Proteins [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(6): 2600-2612. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||