





Acta Veterinaria et Zootechnica Sinica ›› 2025, Vol. 56 ›› Issue (5): 2157-2167.doi: 10.11843/j.issn.0366-6964.2025.05.016
• Animal Genetics and Breeding • Previous Articles Next Articles
QIAO Liying1,2( ), WANG Wannian1, ZHANG Li1, PANG Zhixu1, ZHANG Siying1, LI Yifan1, LIU Wenzhong1,2,*(
), WANG Wannian1, ZHANG Li1, PANG Zhixu1, ZHANG Siying1, LI Yifan1, LIU Wenzhong1,2,*( )
)
Received:2024-10-24
Online:2025-05-23
Published:2025-05-27
Contact:
LIU Wenzhong
E-mail:liyingqiao1970@163.com;tglwzyc@163.com
CLC Number:
QIAO Liying, WANG Wannian, ZHANG Li, PANG Zhixu, ZHANG Siying, LI Yifan, LIU Wenzhong. Machine Learning Methods for Sheep Breed Classification Based on Genomic Markers[J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(5): 2157-2167.
Table 1
Breeds and number of individuals retained after quality control"
| 品种 Breed | 个体数 Number of individuals |
| 芬兰羊Finnish sheep(Finn) | 54 |
| 冰岛羊Icelandic sheep(Icelandic) | 54 |
| 罗曼诺夫羊Romanov sheep(Romanov) | 79 |
| 特塞尔羊Texel sheep(Texel) | 59 |
| 多浪羊Duolang sheep(DL) | 119 |
| 湖羊Hu sheep(Hu) | 112 |
| 大尾寒羊Large Tail Han sheep(LTH) | 106 |
| 泗水裘皮羊Sishui Fur sheep(SSF) | 58 |
| 小尾寒羊Small Tail Han sheep(STH) | 102 |
| 洼地羊Wadi sheep(WD) | 146 |
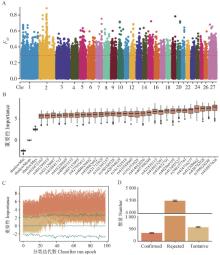
Fig. 3
SNPs screening process and results A.A total of 5 361 SNPs were screened by FST analysis; B. Importance scores of the first 30 SNPs in the Boruta algorithm at each iteration; C. Importance scores of SNPs during multiple iterations; D. Number and standard deviation of SNPs marked as confirmed, rejected and tentative in 10 replicates"
| 1 |
GREENER J G , KANDATHIL S M , MOFFAT L , et al. A guide to machine learning for biologists[J]. Nat Rev Mol Cell Biol, 2022, 23 (1): 40- 55.
doi: 10.1038/s41580-021-00407-0 |
| 2 |
CHAFAI N , HAYAH I , HOUAGA I , et al. A review of machine learning models applied to genomic prediction in animal breeding[J]. Front Genet, 2023, 14, 1150596.
doi: 10.3389/fgene.2023.1150596 |
| 3 |
李棉燕, 王立贤, 赵福平. 机器学习在动物基因组选择中的研究进展[J]. 中国农业科学, 2023, 56 (18): 3682- 3692.
doi: 10.3864/j.issn.0578-1752.2023.18.015 |
|
LI M Y , WANG L X , ZHAO F P . Research progress on machine learning for genomic selection in animals[J]. Scientia Agricultura Sinica, 2023, 56 (18): 3682- 3692.
doi: 10.3864/j.issn.0578-1752.2023.18.015 |
|
| 4 |
LIU R , XU Z , TENG J , et al. Evaluation of six machine learning classification algorithms in pig breed identification using SNPs array data[J]. Anim Genet, 2023, 54 (2): 113- 122.
doi: 10.1111/age.13279 |
| 5 |
ZHAO C , WANG D , YANG C , et al. Population structure and breed identification of Chinese indigenous sheep breeds using whole genome SNPs and InDels[J]. Genet Sel Evol, 2024, 56 (1): 60.
doi: 10.1186/s12711-024-00927-1 |
| 6 |
JIE W , LEI Q X , CAO D G , et al. Whole genome SNPs among 8 chicken breeds enable identification of genetic signatures that underlie breed features[J]. J Integr Agr, 2023, 22 (7): 2200- 2212.
doi: 10.1016/j.jia.2022.11.007 |
| 7 |
LIAKOS K G , BUSATO P , MOSHOU D , et al. Machine learning in agriculture: A review[J]. Sensors, 2018, 18 (8): 2674.
doi: 10.3390/s18082674 |
| 8 |
AYO F E , AWOTUNDE J B , FOLORUNSO S O , et al. A genomic rule-based KNN model for fast flux botnet detection[J]. Egypt Inform J, 2023, 24 (2): 313- 325.
doi: 10.1016/j.eij.2023.05.002 |
| 9 |
YUAN Y , SHI C , ZHAO H . Machine learning-enabled genome mining and bioactivity prediction of natural products[J]. ACS Synth Biol, 2023, 12 (9): 2650- 2662.
doi: 10.1021/acssynbio.3c00234 |
| 10 |
PEIGNIER S , SORIN B , CALEVRO F . Ensemble learning based gene regulatory network inference[J]. Int J Artif Intell T, 2023, 32 (5): 2360005.
doi: 10.1142/S0218213023600059 |
| 11 |
GOUDET J , WEIR B S . An allele-sharing, moment-based estimator of global, population-specific and population-pair F ST under a general model of population structure[J]. PLoS Genet, 2023, 19 (11): e1010871.
doi: 10.1371/journal.pgen.1010871 |
| 12 |
ZHOU H , XIN Y , LI S . A diabetes prediction model based on Boruta feature selection and ensemble learning[J]. BMC Bioinformatics, 2023, 24 (1): 224.
doi: 10.1186/s12859-023-05300-5 |
| 13 | 梁卉, 王雪, 司敬方, 等. 利用基因组标记和机器学习算法对中国牛品种的分类准确性研究[J]. 遗传, 2024, 46 (7): 530- 539. |
| LIANG H , WANG X , SI J F , et al. Classification accuracy of machine learning algorithms for Chinese local cattle breeds using genomic markers[J]. Hereditas (Beijing), 2024, 46 (7): 530- 539. | |
| 14 |
RAMÍREZ-GALLEGO S , LASTRA I , MARTÍNEZ-REGO D , et al. Fast-mRMR: Fast minimum redundancy maximum relevance algorithm for high-dimensional big data[J]. Int J Intell Syst, 2017, 32 (2): 134- 152.
doi: 10.1002/int.21833 |
| 15 |
LI X , LI H , YANG Z , et al. Distribution rules of 8-mer spectra and characterization of evolution state in animal genome sequences[J]. BMC Genomics, 2024, 25 (1): 855.
doi: 10.1186/s12864-024-10786-1 |
| 16 |
SCHIAVO G , BERTOLINI F , GALIMBERTI G , et al. A machine learning approach for the identification of population-informative markers from high-throughput genotyping data: application to several pig breeds[J]. Animal, 2020, 14 (2): 223- 232.
doi: 10.1017/S1751731119002167 |
| 17 | DAN S , MANDAL S N , GHOSH P , et al. Principal component analysis in pig breeds identification[J]. Indian J Anim Sci, 2023, 93 (4): 401- 405. |
| 18 |
PILES M , BERGSMA R , GIANOLA D , et al. Feature selection stability and accuracy of prediction models for genomic prediction of residual feed intake in pigs using machine learning[J]. Front Genet, 2021, 12, 611506.
doi: 10.3389/fgene.2021.611506 |
| 19 |
王万年, 陈思佳, 郜金荣, 等. 基于多层感知机的绵羊限性性状基因组选择模拟研究[J]. 畜牧兽医学报, 2023, 54 (7): 2824- 2835.
doi: 10.11843/j.issn.0366-6964.2023.07.015 |
|
WANG W N , CHEN S J , GAO J R , et al. Simulation study on genomic selection of sex-limited traits using multilayer perceptron in sheep[J]. Acta Veterinaria et Zootechnica Sinica, 2023, 54 (7): 2824- 2835.
doi: 10.11843/j.issn.0366-6964.2023.07.015 |
|
| 20 |
MORONI B , BRAMBILLA A , ROSSI L , et al. Hybridization between Alpine Ibex and Domestic Goat in the Alps: A Sporadic and Localized Phenomenon?[J]. Animals, 2022, 12 (6): 751.
doi: 10.3390/ani12060751 |
| 21 |
WANG Z H , ZHU Q H , LI X , et al. iSheep: an integrated resource for sheep genome, variant and phenotype[J]. Front Genet, 2021, 12, 714852.
doi: 10.3389/fgene.2021.714852 |
| 22 |
PURCELL S , NEALE B , TODD-BROWN K , et al. PLINK: a tool set for whole-genome association and population-based linkage analyses[J]. Am J Hum Genet, 2007, 81 (3): 559- 575.
doi: 10.1086/519795 |
| 23 |
IHAKA R , GENTLEMAN R . R: a language for data analysis and graphics[J]. J Comput Graph Stat, 1996, 5 (3): 299- 314.
doi: 10.1080/10618600.1996.10474713 |
| 24 |
ALEXANDER D H , NOVEMBRE J , LANGE K . Fast model-based estimation of ancestry in unrelated individuals[J]. Genome Res, 2009, 19 (9): 1655- 1664.
doi: 10.1101/gr.094052.109 |
| 25 |
WICKHAM H . ggplot2[J]. Wiley Interdiscip Rev Comput Stat, 2011, 3 (2): 180- 185.
doi: 10.1002/wics.147 |
| 26 | KARATZOGLOU A , SMOLA A , HORNIK K , et al. kernlab-an S4 package for kernel methods in R[J]. J Stat Softw, 2004, 11, 1- 20. |
| 27 | HALDAR A , PAL P , GHOSH S , et al. Body weight prediction using recursive partitioning and regression trees (RPART) model in indian black Bengal goat breed: A machine learning approach[J]. Indian J Anim Res, 2023, 57 (9): 1251- 1257. |
| 28 |
HENGL T , MENDES DE JESUS J , HEUVELINK G B , et al. SoilGrids250m: Global gridded soil information based on machine learning[J]. PLoS One, 2017, 12 (2): e0169748.
doi: 10.1371/journal.pone.0169748 |
| 29 | MEYER D , WIEN F T . Support vector machines[J]. R News, 2001, 1 (3): 23- 26. |
| 30 | RCOLORBREWER S , LIAW M A . Package 'randomforest '[J]. UC Berkeley: Berkeley, CA, USA, 2018, |
| 31 | ALFARO E , GAMEZ M , GARCIA N . Adabag: An R package for classification with boosting and bagging[J]. J Stat Softw, 2013, 54, 1- 35. |
| 32 | RIDGEWAY G . Generalized boosted models: A guide to the gbm package[J]. Update, 2007, 1 (1): 2007. |
| 33 | CHEN T , HE T , BENESTY M , et al. Package 'xgboost '[J]. R Version, 2019, 90 (1-66): 40. |
| 34 |
ROBIN X , TURCK N , HAINARD A , et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves[J]. BMC Bioinformatics, 2011, 12, 77.
doi: 10.1186/1471-2105-12-77 |
| 35 | TAN K , WANG R , LI M , et al. Discriminating soybean seed varieties using hyperspectral imaging and machine learning[J]. J Comput Methods Sci, 2019, 19 (4): 1001- 1015. |
| 36 |
MEADOWS J R S , HIENDLEDER S , KIJAS J W . Haplogroup relationships between domestic and wild sheep resolved using a mitogenome panel[J]. Heredity, 2011, 106 (4): 700- 706.
doi: 10.1038/hdy.2010.122 |
| 37 |
BRAGA-NETO U M , ZOLLANVARI A , DOUGHERTY E R . Cross-validation under separate sampling: strong bias and how to correct it[J]. Bioinformatics, 2014, 30 (23): 3349- 3355.
doi: 10.1093/bioinformatics/btu527 |
| 38 |
LIU R , XU Z , TENG J , et al. Evaluation of six machine learning classification algorithms in pig breed identification using SNPs array data[J]. Anim Genet, 2023, 54 (2): 113- 122.
doi: 10.1111/age.13279 |
| 39 | ZHANG Y , DING C , LI T . Gene selection algorithm by combining reliefF and mRMR[J]. BMC Genomics, 2008, 9 (Suppl 2): 527. |
| 40 |
WANG X , REN J , REN H , et al. Diabetes mellitus early warning and factor analysis using ensemble Bayesian networks with SMOTE-ENN and Boruta[J]. Sci Rep, 2023, 13 (1): 12718.
doi: 10.1038/s41598-023-40036-5 |
| 41 |
AL-MAMUN H A , DANILEVICZ M F , MARSH J I , et al. Exploring genomic feature selection: A comparative analysis of GWAS and machine learning algorithms in a large-scale soybean dataset[J]. Plant Genome, 2025, 18 (1): e20503.
doi: 10.1002/tpg2.20503 |
| 42 |
SARDER M A , MANIRUZZAMAN M , AHAMMED B . Feature selection and classification of leukemia cancer using machine learning techniques[J]. J Mach Learn Res, 2020, 5 (2): 18.
doi: 10.11648/j.mlr.20200502.11 |
| 43 | CHANDRA M A , BEDI S S . Survey on SVM and their application in image classification[J]. J Inf Technol, 2021, 13 (5): 1- 11. |
| 44 |
BLANQUERO R , CARRIZOSA E , RAMÍREZ-COBO P , et al. Variable selection for Naïve Bayes classification[J]. Comput Oper Res, 2021, 135, 105456.
doi: 10.1016/j.cor.2021.105456 |
| 45 |
IMRAN M , BHATTI A , KING D M , et al. Supervised Machine Learning-Based Decision Support for Signal Validation Classification[J]. Drug Saf, 2022, 45 (5): 583- 596.
doi: 10.1007/s40264-022-01159-2 |
| 46 | ZHANG S . Challenges in KNN classification[J]. IEEE Trans Knowl Data Eng, 2021, 34 (10): 4663- 4675. |
| 47 | XU Z , DIAO S , TENG J , et al. Breed identification of meat using machine learning and breed tag SNPs[J]. Food Control, 2021, 125 (1): 107971. |
| 48 |
ZHAO C , WANG D , TENG J , et al. Breed identification using breed-informative SNPs and machine learning based on whole genome sequence data and SNP chip data[J]. J Anim Sci Biotechnol, 2023, 14 (1): 85.
doi: 10.1186/s40104-023-00880-x |
| 49 |
NEETHIRAJAN S . Affective state recognition in livestock-artificial intelligence approaches[J]. Animals, 2022, 12 (6): 759.
doi: 10.3390/ani12060759 |
| [1] | SUN Guoxin, LI Yunhua, SAI Yin, GUO Wenhua, ZHAO Yanhong, ZHANG Manxin, LIU Jiasen. Population Structure Analysis and Economic Traits Related Selection Signal Detection of Hu Sheep [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(5): 2168-2181. |
| [2] | LI Xiaowei, TIAN Wei, LIU Yuan, LI Huixia. Study on the Difference of m6A Methylation Modification in Ovarian Granulosa Cells of Hu Sheep under Heat Stress [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(4): 1712-1721. |
| [3] | MA Yingtian, JIANG Luyao, LI Zengkai, QIN Jianping, ZHAO Jianhua, HE Yufang, SONG Yuxuan, ZHANG Lei. Effect of Cyanidin-3-rutinoside on Cryopreservation of Semen of Dairy Sheep [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(4): 1768-1778. |
| [4] | YANG Yang, LI Liangyuan, WAN Pengcheng, LU Shouliang, LIU Changbin, YANG Hua, WANG Limin, DAI Rong, ZHOU Ping. Screening and Analysis of Core Genes and Key lncRNAs for Seasonal Estrus Traits in Sheep [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(3): 1264-1277. |
| [5] | YANG Miaomiao, XIE Li, JIAN Baoyi, LUO Chaowei, XIE Zhuojun, ZHU Piao, ZHOU Tianri, LI Hua, XIANG Hai. Construction and Optimization of Prediction Models for Abdominal Fat Deposition in Adult Hens based on Early Body Size Traits using Machine Learning [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(2): 548-558. |
| [6] | XI Haijiao, LI Jinquan, ZHANG Yanjun, WANG Ruijun, LÜ Qi, MEI Bujun, WANG Na, SU Rui, WANG Zhiying. Influence of Dominance Effects on the Accuracy of Breeding Value Estimation of Cashmere Production and Cashmere Diameter in Inner Mongolia Cashmere Goats [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(2): 571-581. |
| [7] | HE Yu, WANG Xiangyu, DI Ran, CHU Mingxing, LIANG Chen. BMP4/SMAD4 Downregulates GJA1 Gene Expression to Affect the Gap Junctional Intercellular Communication Activity in Sheep Ovarian Granulosa Cells [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(2): 679-688. |
| [8] | CHU Yijian, CUI Jiuzeng, LI Zengkai, ZHANG Lei, CHU Tingting, HUANG Yanping, SONG Yuxuan. Comparative Study on Vaginal Microorganisms in Pre-endometrial Receptivity and Endometrial Receptivity of Sheep [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(2): 689-699. |
| [9] | WANG Xiaofei, WANG Bosen, WEI Mengyao, JIANG Luyao, XU Ganggang, LIU Jiaxin, MA Yingtian, WANG Li, SONG Yuxuan, ZHANG Lei. Study on the Role of Ewe's Milk in Ameliorating Pathological Changes in the Liver and Kidney of Mice in a Diabetes Model [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(2): 870-882. |
| [10] | YANG Wenpan, LIU Xiangjie, LUO Dongxiang, CHEN Menghui, XIE Ying, FANG Yuexin, LIN Tingyan, LI Aimin, LI Wenjing, DENG Zheng, DING Nengshui. Research on Genomic Selection of Reproductive Traits in Landrace Pigs Based on Chip Data [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(1): 213-221. |
| [11] | LI Wei, WU Xilong, ZHAO Xingrui, XU Lanjiao, YANG Xiaobin, SONG Xiaozhen. Effects of Chinese Medicine Jianpisiwei Formulas on Growth Performance, Rumen Fermentation and Microbiota Composition of Weaned Hu Sheep [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(1): 466-478. |
| [12] | Yuhang JIA, Liangfu GUO, Runan ZHANG, Ayong ZHAO, Yufang LIU, Mingxing CHU. miR-127 Regulated the Proliferation and Differentiation of Sheep Skeletal Myoblasts and Its Transcription Factor PAX3 Screening [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(9): 3864-3875. |
| [13] | Yiming GONG, Yixuan JIA, Jiajun LI, Xiangyu WANG, Xiaoyun HE, Mingxing CHU, Ran DI. BMP/SMAD Pathway Activity and Protein Expression Profiles in Ovarian Follicles with Different Diameters in Diverse FecB Genotyped Ewes [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(9): 3957-3967. |
| [14] | Peng SHEN, Yi WANG, Weijie REN, Yongchun YANG, Houhui SONG, Zhiliang WANG. Meta Analysis of Immune Antibody Monitoring for Lumpy Skin Disease [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(8): 3649-3658. |
| [15] | Jinbu WANG, Jia LI, Deming REN, Lixian WANG, Ligang WANG. Progress in the Application of Machine Learning in Livestock and Poultry Genomic Selection [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(7): 2775-2785. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||