





Acta Veterinaria et Zootechnica Sinica ›› 2025, Vol. 56 ›› Issue (8): 3787-3801.doi: 10.11843/j.issn.0366-6964.2025.08.020
• Animal Genetics and Breeding • Previous Articles Next Articles
REN Qianzi1( ), ZHANG Baizhong2(
), ZHANG Baizhong2( ), WANG Zhenqing1, WANG Xianglin3, GONG Ying1, HU Renke2, PU Yabin1, SU Peng1, LI Yefang1, MA Yuehui1, LI Haobang2,*(
), WANG Zhenqing1, WANG Xianglin3, GONG Ying1, HU Renke2, PU Yabin1, SU Peng1, LI Yefang1, MA Yuehui1, LI Haobang2,*( ), JIANG Lin1,*(
), JIANG Lin1,*( )
)
Received:2025-01-15
Online:2025-08-23
Published:2025-08-28
Contact:
LI Haobang, JIANG Lin
E-mail:renqianzi677@gmail.com;705048333@qq.com;lhb.m2002@163.com;jianglin@caas.cn
CLC Number:
REN Qianzi, ZHANG Baizhong, WANG Zhenqing, WANG Xianglin, GONG Ying, HU Renke, PU Yabin, SU Peng, LI Yefang, MA Yuehui, LI Haobang, JIANG Lin. Genetic Evolutionary Analysis of Wuxue Goat Based on Whole Genome Resequencing[J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(8): 3787-3801.
Table 1
Samples information for the 6 goat populations"
| 群体Group | 英文名称English name | 数量Number | 样品来源Source |
| 武雪山羊WXG | Wuxue goat | 18 | 湖南省湘西土家族苗族自治州 |
| 湘东黑山羊XDB | Xiangdong Black goat | 9 | 湖南省浏阳市 |
| 合川白山羊HCW | Hechuan White goat | 9 | 重庆市合川县 |
| 大足黑山羊DZB | Dazu Black goat | 9 | 重庆市大足县 |
| 云岭山羊YLG | Yunling goat | 10 | 云南省楚雄彝族自治州 |
| 马关无角山羊MGG | Maguan Poll goat | 11 | 云南省文山壮族苗族自治州马关县 |
Table 2
Annotation information of all SNPs"
| 类型Type (alphabetical order) | 数量Count | 占比/% Percent |
| DOWNSTREAM | 2 135 800 | 5.031 |
| EXON | 270 736 | 0.638 |
| INTERGENIC | 9 102 451 | 21.440 |
| INTRON | 14 166 150 | 33.367 |
| SPLICE_SITE_ACCEPTOR | 239 | 0.001 |
| SPLICE_SITE_DONOR | 252 | 0.001 |
| SPLICE_SITE_REGION | 22 952 | 0.054 |
| TRANSCRIPT | 14 442 290 | 34.018 |
| UPSTREAM | 2 107 844 | 4.965 |
| UTR_3_PRIME | 164 145 | 0.387 |
| UTR_5_PRIME | 42 447 | 0.100 |
| 3_prime_UTR_variant | 164 145 | 0.386 |
| 5_prime_UTR_premature_start_codon_gain_variant | 6 098 | 0.014 |
| 5_prime_UTR_variant | 36 349 | 0.086 |
| downstream_gene_variant | 2 135 800 | 5.028 |
| initiator_codon_variant | 9 | 0.000 |
| intergenic_region | 9 102 451 | 21.428 |
| intragenic_variant | 168 001 | 0.395 |
| intron_variant | 14 185 337 | 33.394 |
| missense_variant | 69 583 | 0.164 |
| non_coding_transcript_exon_variant | 53 867 | 0.127 |
| non_coding_transcript_variant | 14 274 289 | 33.603 |
| splice_acceptor_variant | 239 | 0.001 |
| splice_donor_variant | 263 | 0.001 |
| splice_region_variant | 24 149 | 0.057 |
| start_lost | 78 | 0.000 |
| start_retained_variant | 30 | 0.000 |
| stop_gained | 648 | 0.002 |
| stop_lost | 92 | 0.000 |
| stop_retained_variant | 127 | 0.000 |
| synonymous_variant | 149 823 | 0.353 |
| upstream_gene_variant | 2 107 844 | 4.962 |
Table 3
Genetic diversity of 6 goat populations"
| 品种 Breed | 核苷酸多态性 π(mean) | 观测杂合度 Observed heterozygosity(Ho) | 期望杂合度 Expected heterozygosity(He) |
| 大足黑山羊DZB | 0.001 671 572 | 0.253 802 | 0.272 038 |
| 合川白山羊HCW | 0.001 481 751 | 0.242 553 | 0.242 862 |
| 马关无角山羊MGG | 0.001 706 207 | 0.296 721 | 0.275 427 |
| 武雪山羊WXG | 0.001 585 495 | 0.235 027 | 0.261 443 |
| 湘东黑山羊XDB | 0.001 637 571 | 0.265 953 | 0.264 043 |
| 云岭山羊YLG | 0.001 523 224 | 0.238 900 | 0.250 394 |
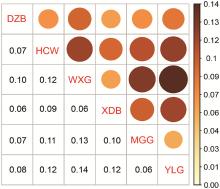
Fig. 10
Goat population differentiation index FST The different colours and sizes of the circles in the figure represent the size of the FST value, the larger the FST value, the darker the colour and the larger the size; the smaller the FST value, the lighter the colour and the smaller the size"
| 1 |
姚华清,田斌,秦茂.浅析龙山县武雪山羊遗传资源保护与利用[J].湖南畜牧兽医,2023(2):1-3.
doi: 10.3969/j.issn.1006-4907.2023.02.001 |
|
YAOH Q,TIANB,QINM.Conservation and utilisation of genetic resources of Wuxue goats in Longshan County[J].Hunan Animal Husbandry and Veterinary Medicine,2023(2):1-3.
doi: 10.3969/j.issn.1006-4907.2023.02.001 |
|
| 2 |
石琦,杨建昌.武雪山羊[J].湖南农业,2022(6):29.
doi: 10.3969/j.issn.1005-362X.2022.06.027 |
|
SHIQ,YANGJ C.Wuxue goats[J].Hunan Agriculture,2022(6):29.
doi: 10.3969/j.issn.1005-362X.2022.06.027 |
|
| 3 | 林泽榜.武雪山羊林间养殖发展现状及思路[J].基层农技推广,2013,1(11):59-62. |
| LINZ B.Current situation and ideas for the development of forest farming of Wuxue goats[J].Grassroots Agricultural Extension,2013,1(11):59-62. | |
| 4 |
IAMARTINOD,BRUZZONEA,LANZAA,et al.Genetic diversity of Southern Italian goat populations assessed by microsatellite markers[J].Small Ruminant Res,2005,57(2-3):249-255.
doi: 10.1016/j.smallrumres.2004.08.003 |
| 5 |
DALYK G,MATTIANGELIV,HAREA J,et al.Herded and hunted goat genomes from the dawn of domestication in the Zagros Mountains[J].Proc Natl Acad Sci,2021,118(25):e2100901118.
doi: 10.1073/pnas.2100901118 |
| 6 |
HANOTTEO,BRADLEYD G,OCHIENGJ W,et al.African pastoralism: genetic imprints of origins and migrations[J].Science,2002,296(5566):336-339.
doi: 10.1126/science.1069878 |
| 7 |
LVF H,CAOY H,LIUG J,et al.Whole-genome resequencing of worldwide wild and domestic sheep elucidates genetic diversity, introgression, and agronomically important loci[J].Mol Biol Evol,2022,39(2):msab353.
doi: 10.1093/molbev/msab353 |
| 8 |
CHENN B,CAIY D,CHENQ M,et al.Whole-genome resequencing reveals world-wide ancestry and adaptive introgression events of domesticated cattle in East Asia[J].Nat Commun,2018,9(1):2337.
doi: 10.1038/s41467-018-04737-0 |
| 9 |
BRUNO-DE-SOUSAC,MARTINEZA,GINJAC,et al.Genetic diversity and population structure in Portuguese goat breeds[J].Livest Sci,2011,135(2-3):131-139.
doi: 10.1016/j.livsci.2010.06.159 |
| 10 |
SENCZUKG,MACRÌM,DI CIVITAM,et al.The demographic history and adaptation of Canarian goat breeds to environmental conditions through the use of genome-wide SNP data[J].Genet Select Evol,2024,56(1):2.
doi: 10.1186/s12711-023-00869-0 |
| 11 |
YANGJ,WANGD F,HUANGJ H,et al.Structural variant landscapes reveal convergent signatures of evolution in sheep and goats[J].Genome Biol,2024,25(1):148.
doi: 10.1186/s13059-024-03288-6 |
| 12 |
CHANGL,ZHENGY,LIS,et al.Identification of genomic characteristics and selective signals in Guizhou black goat[J].BMC Genomics,2024,25(1):164.
doi: 10.1186/s12864-023-09954-6 |
| 13 |
ZHENGZ Q,WANGX H,LIM,et al.The origin of domestication genes in goats[J].Sci Adv,2020,6(21):eaaz5216.
doi: 10.1126/sciadv.aaz5216 |
| 14 |
LEIC Z,CHENH,ZHANGH C,et al.Origin and phylogeographical structure of Chinese cattle[J].Anim Genet,2006,37(6):579-582.
doi: 10.1111/j.1365-2052.2006.01524.x |
| 15 |
BAH,JIAB,WANGG,et al.Genome-wide SNP discovery and analysis of genetic diversity in farmed sika deer (Cervus nippon) in northeast China using double-digest restriction site-associated DNA sequencing[J].G3: Genes, Genomes, Genetics,2017,7(9):3169-3176.
doi: 10.1534/g3.117.300082 |
| 16 |
刘珍妮,李建军,连海,等.基于全基因组重测序对赣州番鸭的群体进化分析[J].畜牧兽医学报,2024,55(11):4992-5002.
doi: 10.11843/j.issn.0366-6964.2024.11.018 |
|
LIUZ N,LIJ J,LIANH,et al.Population evolution analysis of ganzhou muscovy duck based on whole genome resequencing[J].Acta Veterinaria et Zootechnica Sinica,2024,55(11):4992-5002.
doi: 10.11843/j.issn.0366-6964.2024.11.018 |
|
| 17 |
SAITBEKOVAN,GAILLARDC,OBEXER-RUFFG,et al.Genetic diversity in Swiss goat breeds based on microsatellite analysis[J].Anim Genet,1999,30(1):36-41.
doi: 10.1046/j.1365-2052.1999.00429.x |
| 18 |
CHENS F,ZHOUY Q,CHENY R,et al.fastp: an ultra-fast all-in-one FASTQ preprocessor[J].Bioinformatics,2018,34(17):i884-i890.
doi: 10.1093/bioinformatics/bty560 |
| 19 |
LIH,DURBINR.Fast and accurate short read alignment with Burrows-Wheeler transform[J].Bioinformatics,2009,25(14):1754-1760.
doi: 10.1093/bioinformatics/btp324 |
| 20 |
BICKHARTD M,ROSENB D,KORENS,et al.Single-molecule sequencing and chromatin conformation capture enable de novo reference assembly of the domestic goat genome[J].Nat Genet,2017,49(4):643-650.
doi: 10.1038/ng.3802 |
| 21 |
LIH,HANDSAKERB,WYSOKERA,et al.The sequence alignment/map format and SAMtools[J].Bioinformatics,2009,25(16):2078-2079.
doi: 10.1093/bioinformatics/btp352 |
| 22 |
MCKENNAA,HANNAM,BANKSE,et al.The genome analysis toolkit: a mapReduce framework for analyzing next-generation DNA sequencing data[J].Genome Research,2010,20(9):1297-1303.
doi: 10.1101/gr.107524.110 |
| 23 |
CINGOLANIP,PLATTSA,WANG LEL,et al.A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3[J].Fly (Austin),2012,6(2):80-92.
doi: 10.4161/fly.19695 |
| 24 | CHANGC C,CHOWC C,TELLIERL C,et al.Second-generation PLINK: rising to the challenge of larger and richer datasets[J].Gigascience,2015,4(1):s13742-13015-10047-13748. |
| 25 |
YANGJ,LEES H,GODDARDM E,et al.GCTA: a tool for genome-wide complex trait analysis[J].Am J Hum Genet,2011,88(1):76-82.
doi: 10.1016/j.ajhg.2010.11.011 |
| 26 | RACINEJ S.RStudio: a platform-independent IDE for R and Sweave[J].J Appl Econom,2012(1):167-172. |
| 27 |
TAMURAK,STECHERG,KUMARS.MEGA11: molecular evolutionary genetics analysis version 11[J].Mol Biol Evol,2021,38(7):3022-3027.
doi: 10.1093/molbev/msab120 |
| 28 | TREESR P.The neighbor-joining method: a new method for[J].Mol Biol Evol,1987,4(4):406-425. |
| 29 |
PRITCHARDJ K,STEPHENSM,DONNELLYP.Inference of population structure using multilocus genotype data[J].Genetics,2000,155(2):945-959.
doi: 10.1093/genetics/155.2.945 |
| 30 |
LAWSOND J,VAN DORPL,FALUSHD.A tutorial on how not to over-interpret STRUCTURE and ADMIXTURE bar plots[J].Nat Commun,2018,9(1):3258.
doi: 10.1038/s41467-018-05257-7 |
| 31 |
GOODALL-COPESTAKEW,TARLINGG,MURPHYE.On the comparison of population-level estimates of haplotype and nucleotide diversity: a case study using the gene cox1 in animals[J].Heredity,2012,109(1):50-56.
doi: 10.1038/hdy.2012.12 |
| 32 |
CEBALLOSF C,JOSHIP K,CLARKD W,et al.Runs of homozygosity: windows into population history and trait architecture[J].Nat Rev Genet,2018,19(4):220-234.
doi: 10.1038/nrg.2017.109 |
| 33 | WEIRB S,COCKERHAMC C.Estimating F-statistics for the analysis of population structure[J].Evolution,1984,38(6):1358-1370. |
| 34 |
HILLW,ROBERTSONA.Linkage disequilibrium in finite populations[J].Theor Appl Genet,1968,38,226-231.
doi: 10.1007/BF01245622 |
| 35 |
ZHANGC,DONGS S,XUJ Y,et al.PopLDdecay: a fast and effective tool for linkage disequilibrium decay analysis based on variant call format files[J].Bioinformatics,2019,35(10):1786-1788.
doi: 10.1093/bioinformatics/bty875 |
| 36 |
王浩宇,马克岩,李讨讨,等.基于简化基因组测序评估小骨山羊群体遗传多样性和群体结构[J].畜牧兽医学报,2025,56(3):1170-1179.
doi: 10.11843/j.issn.0366-6964.2025.03.018 |
|
WANGH Y,MAK Y,LIT T,et al.Population genetic diversity and population structure analysis of small-boned goat based on specific-locus amplified fragment sequencing[J].Acta Veterinaria et Zootechnica Sinica,2025,56(3):1170-1179.
doi: 10.11843/j.issn.0366-6964.2025.03.018 |
|
| 37 |
胡紫平,王立刚,宗文成,等.基于基因组SNP和ROH的剑白香猪群体遗传结构解析[J].畜牧兽医学报,2023,54(10):4117-4125.
doi: 10.11843/j.issn.0366-6964.2023.10.011 |
|
HUZ P,WANGL G,ZONGW C,et al.Genetic structure analysis of Jianbai Xiang Pig population based on genomic SNP and ROH[J].Acta Veterinaria et Zootechnica Sinica,2023,54(10):4117-4125.
doi: 10.11843/j.issn.0366-6964.2023.10.011 |
|
| 38 | 余道宁,王佟,马晓明,等.西藏牦牛群体遗传多样性及遗传结构分析[J].中国草食动物科学,2024,44(2):39-43. |
| YUD N,WANGT,MAX M,et al.Study on genetic diversity and genetic structure of Tibetan yak populations[J].Chinese Herbivore Science,2024,44(2):39-43. | |
| 39 | 王心彤,张艳丽,龚颖,等.基于世界家马群体遗传结构构建藏马特异性遗传标签[J].中国畜牧兽医,2025,52(1):238-248. |
| WANGX T,ZHANGY L,GONGY,et al.Construction of specific genetic markers for tibetan horses through population structure analysis of global domestic horses[J].China Animal Husbandry & Veterinary Medicine,2025,52(1):238-248. | |
| 40 |
徐扩卫,李卓辉,冷堂健,等.基于全基因组重测序SNP分析宁蒗高原鸡保种群的群体遗传多样性和群体遗传结构[J].畜牧兽医学报,2024,55(12):5498-5510.
doi: 10.11843/j.issn.0366-6964.2024.12.016 |
|
XUK W,LIZ H,LENGT J,et al.Analysis of population genetic diversity and population genetic structure of conservation population in ninglang plateau chickens based on whole-genome resequencing SNP[J].Acta Veterinaria et Zootechnica Sinica,2024,55(12):5498-5510.
doi: 10.11843/j.issn.0366-6964.2024.12.016 |
|
| 41 |
RINGNÉRM.What is principal component analysis?[J].Nat Biotechnol,2008,26(3):303-304.
doi: 10.1038/nbt0308-303 |
| 42 |
NOVEMBREJ,STEPHENSM.Interpreting principal component analyses of spatial population genetic variation[J].Nat Genet,2008,40(5):646-649.
doi: 10.1038/ng.139 |
| 43 |
MENOZZIP,PIAZZAA,CAVALLI-SFORZAL.Synthetic maps of human gene frequencies in Europeans: These maps indicate that early farmers of the Near East spread to all of Europe in the Neolithic[J].Science,1978,201(4358):786-792.
doi: 10.1126/science.356262 |
| 44 |
TAMURAK,NEIM,KUMARS.Prospects for inferring very large phylogenies by using the neighbor-joining method[J].Proc Natl Acad Sci,2004,101(30):11030-11035.
doi: 10.1073/pnas.0404206101 |
| 45 |
杨章平,毛永江,马月辉,等.中国南方地区7个山羊群体的遗传分化与基因流分析[J].畜牧兽医学报,2008,39(5):562-569.
doi: 10.3321/j.issn:0366-6964.2008.05.006 |
|
YANGZ P,MAOY J,MAY H,et al.Genetic differentiation and gene flow among seven indigenous goat populations in South of China[J].Acta Veterinaria et Zootechnica Sinica,2008,39(5):562-569.
doi: 10.3321/j.issn:0366-6964.2008.05.006 |
|
| 46 |
LIC,WANGX,LIH,et al.Whole-genome resequencing reveals diversity and selective signals in the Wuxue goat[J].Anim Genet,2024,55(4):575-587.
doi: 10.1111/age.13437 |
| 47 | 毛永江. 中国牛亚科家畜六个群体遗传多样性与遗传分化及其统计方法的研究[D]. 扬州: 扬州大学, 2006. |
| MAO Y J. The genetic diversity, genetic differentiation of six cattle populations in bovidae in China and the statistical methods of genetic diversity research[D]. Yangzhou: Yangzhou University, 2006. (in Chinese) | |
| 48 |
HANSSONB,WESTERBERGL.On the correlation between heterozygosity and fitness in natural populations[J].Mol Ecol,2002,11(12):2467-2474.
doi: 10.1046/j.1365-294X.2002.01644.x |
| 49 | 李超杰,蒙萌,李博,等.利用微卫星标记分析山西3个地方牛群体的遗传多样性和遗传分化[J].中国畜牧兽医,2025,52(1):226-237. |
| LIC J,MENGM,LIB,et al.Analysis of genetic diversity and genetic differentiation in 3 local cattle populations of shanxi province using microsatellite markers[J].China Animal Husbandry & Veterinary Medicine,2025,52(1):226-237. | |
| 50 | 贾小姣,陈亚乐,王诗佳,等.9个山羊品种微卫星DNA遗传多样性分析[J].安徽农业大学学报,2019,46(5):779-784. |
| JIAX J,CHENY L,WANGS J,et al.Genetic diversity analysis of microsatellite DNA of nine goat breeds[J].Journal of Anhui Agricultural University,2019,46(5):779-784. | |
| 51 |
BERTOLINIF,CARDOSOT F,MARRASG,et al.Genome-wide patterns of homozygosity provide clues about the population history and adaptation of goats[J].Genet Sel Evol,2018,50,1-12.
doi: 10.1186/s12711-018-0374-1 |
| 52 |
MEIRMANSP G,HEDRICKP W.Assessing population structure: FST and related measures[J].Mol Ecol Resources,2011,11(1):5-18.
doi: 10.1111/j.1755-0998.2010.02927.x |
| 53 |
CALDWELLK S,RUSSELLJ,LANGRIDGEP,et al.Extreme population-dependent linkage disequilibrium detected in an inbreeding plant species, Hordeum vulgare[J].Genetics,2006,172(1):557-567.
doi: 10.1534/genetics.104.038489 |
| 54 | GARCÍA-GÁMEZE,SAHANAG,GUTIÉRREZ-GILB,et al.Linkage disequilibrium and inbreeding estimation in Spanish Churra sheep[J].BMC Genet,2012,13,1-11. |
| [1] | LIU Sha, YANG Caichun, ZHANG Xiaoyu, CHEN Qiong, LIU Xiong, CHEN Hongbo, ZHOU Huanhuan, SHI Liangyu. Population Genetic Structure and Genome-wide Runs of Homozygosity Analysis in Meihuaxing Pigs Based on 80K SNP Chip [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(8): 3749-3760. |
| [2] | MIAO Junjie, ZHANG Riquan, WU Houyi, YOU Xinming, HUANG Yiwen, HUANG Xiaoying, GUO Zhenyang, LIU Jianlin, XIAO Weihua, GUO Tianhua, CHEN Hao, KANG Dongliu. Genome-Wide SNP Analysis Revealed the Characteristics of Germplasm Resources and Genetic Diversity of Jinggang Black-Palm Geese [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(7): 3199-3209. |
| [3] | WANG Qinqian, GAO Zhendong, LU Ying, MA Ruoshan, DENG Weidong, HE Xiaoming. Research Progress of Whole Genome Resequencing in Chinese Indigenous Cattle [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(5): 2026-2037. |
| [4] | YAO Tingting, LI Hao, YAN Huixuan, CAO Yifan, Cirengluobu , Suolangquji , Nimacangjue , ZHAO Li, Danzengluosang , Silangwangmu , Basangzhuzha , CHEN Ningbo. Genetic Diversity of Mitochondrial Genome and Maternal Origin of 10 Cattle Populations in Tibet Autonomous Region [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(5): 2194-2202. |
| [5] | WANG Haoyu, MA Keyan, LI Taotao, LI Dengpan, ZHAO Qing, MA Youji. Population Genetic Diversity and Population Structure Analysis of Small-boned Goat Based on Specific-Locus Amplified Fragment Sequencing [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(3): 1170-1179. |
| [6] | HU Xin, YOU Wei, JIANG Fugui, CHENG Haijian, SUN Zhigang, SONG Enliang. Analysis of Genetic Diversity and Population Structure of Simmental Cattle Based on Whole Genome Resequencing [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(3): 1189-1202. |
| [7] | Siyu LIU, Man ZHANG, Yan ZHANG, Zhitong WEI, Xinglei QI, Tengyun GAO, Xian LIU, Dong LIANG, Tong FU. Evaluation of the Conservation Effect in Nanyang Cattle Based on Resequencing Data [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(9): 3876-3886. |
| [8] | Hongyan HUANG, Liyun ZHANG, Zhirong HUANG, Zhongping WU, Xumeng ZHANG, Hongjia OUYANG, Junpeng CHEN, Zhenping LIN, Yunbo TIAN, Xiujin LI, Yunmao HUANG. The Study on Population Genetic Diversity and Genome-wide Association Study of Body Weight and Size Traits for Lion-head Geese [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(9): 3914-3924. |
| [9] | Tao ZHANG, Jiaqi LI, Lei XU, Dan WANG, Menghua ZHANG, Tao ZHANG, Mengjie YAN, Weitao WANG, Shoumin FAN, Xixia HUANG. Detection and Population Structure Analysis of Genomic Structural Variation in Xinjiang Brown Cattle Based on Whole Genome Resequencing Data [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(8): 3427-3435. |
| [10] | SONG Kelin, YAN Zunqiang, WANG Pengfei, CHENG Wenhao, LI Jie, BAI Yaqin, SUN Guohu, GUN Shuangbao. Analysis on Genetic Diversity and Genetic Structure Based on SNP Chips of Huixian Qingni Black Pig [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(3): 995-1006. |
| [11] | REN Yuwei, CHEN Xing, LIN Yanning, HUANG Xiaoxian, HONG Lingling, WANG Feng, SUN Ruiping, ZHANG Yan, LIU Hailong, ZHENG Xinli, CHAO Zhe. Investigating the Influencing Factors of Egg Laying Performance in Wenchang Chickens Based on Whole Genome Resequencing [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(2): 502-514. |
| [12] | CHENG Xinyan, WANG Shiyuan, JI Yebiao, HUANG Sixiu, YANG Jie, MENG Fanming, ZHANG Mao, CAI Gengyuan, LIU Langqing. Evaluation of the Genetic Structure of Conservation Populations of Four Major Local Pig Breeds in Guangdong Province Based on a 50K SNP Chip [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(12): 5464-5477. |
| [13] | XU Kuowei, LI Zhuohui, LENG Tangjian, XIONG Bao, ZHOU Jielong, GUO Panjiang, WANG Yu, CHEN Fenfen. Analysis of Population Genetic Diversity and Population Genetic Structure of Conservation Population in Ninglang Plateau Chickens Based on Whole-genome Resequencing SNP [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(12): 5498-5510. |
| [14] | Huili LIANG, Yujing XIE, Bowen SI, Guiying WANG, Yunliang JIANG, Guiling CAO. Analysis on Genomic Variation and Population Structure of Large-tailed Han Sheep Based on Whole Genome Resequencing [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(11): 4968-4979. |
| [15] | Zhenni LIU, Jianjun LI, Hai LIAN, Xiaowen LEI, Donghai TAN, Qingyuan ZENG, Di CHENG, Yuling TIAN, Zhiwei KONG, Hualiang XIE, Yunping ZHONG. Population Evolution Analysis of Ganzhou Muscovy Duck Based on Whole Genome Resequencing [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(11): 4992-5002. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||