





Acta Veterinaria et Zootechnica Sinica ›› 2025, Vol. 56 ›› Issue (11): 5670-5682.doi: 10.11843/j.issn.0366-6964.2025.11.026
• Preventive Veterinary Medicine • Previous Articles Next Articles
NIE Lianhua1,2( ), FAN Wenyan1,2, LI Mengya1,2, DING Chunhai3, WU Zihao2, WANG Zhihao2, LI Fangfang1,2, JIANG Wei2, HAN Xiangan2,*(
), FAN Wenyan1,2, LI Mengya1,2, DING Chunhai3, WU Zihao2, WANG Zhihao2, LI Fangfang1,2, JIANG Wei2, HAN Xiangan2,*( ), WANG Haidong1,*(
), WANG Haidong1,*( )
)
Received:2025-02-20
Online:2025-11-23
Published:2025-11-27
Contact:
HAN Xiangan, WANG Haidong
E-mail:15110991273@163.com;hanxgan@shvri.ac.cn;wanghaidong@sxau.edu.cn
CLC Number:
NIE Lianhua, FAN Wenyan, LI Mengya, DING Chunhai, WU Zihao, WANG Zhihao, LI Fangfang, JIANG Wei, HAN Xiangan, WANG Haidong. Analysis of Virulence and Antibiotic Resistance Genes in 78 Strains of Staphylococcus aureus from Chicken Arthritis Sources[J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(11): 5670-5682.
Table 1
Primers were used in this study"
| 分类 Classification | 基因名称 Gene name | 引物序列(5′→3′) Primer sequence | 产物大小/bp Length |
| 特异性基因 Specific gene | nuc | GCGATTGATGGTGATACGGTT | 279 |
| AGCCAAGCCTTGACGAACTAAAGC | |||
| 肠毒素 Enterotoxin | seA | GCA GGG AAC AGC TTT AGG C | 521 |
| GTT CTG TAG AAG TAT GAAACA CG | |||
| seC | CTT GTA TGT ATG GAG GAA TAA CAA | 284 | |
| TGC AGG CAT CAT ATC ATA CCAA | |||
| seH | CAA CTG CTG ATT TAG CTC AG | 359 | |
| GTC GAA TGA GTAATC TCT AGG | |||
| seJ | CAT CAG AAC TGT TGT TCC GCT AG | 142 | |
| CTG AAT TTT ACC ATC AAA GGT AC | |||
| 中毒性休克综合征毒素 Toxic shock syndrome toxin | tssT | GCT TGC GAC AAC TGC TAC AG | 559 |
| TGG ATC CGT CAT TCA TTG TTA | |||
| 剥脱毒素 Exfoliative toxin | pvL | ATC ATT AGG TAAAAT GTC TGG ACAT GAT CCA | 433 |
| GCA TCA AST GTA TTG GAT AGC AAAAGC | |||
| etA | CGC TGC GGA CAT TCC TAC ATG G | 676 | |
| TAC ATG CCC GCC ACT TGC TTG T | |||
| etB | CAG ATAAAG AGC TTT ATA CAC ACA TTA | 612 | |
| AGT GAA CTT ATC TTT CTA TTG AAA AAC ACT C | |||
| 溶血素 Hemolysin | hlA | CTG ATT ACT ATC CAA GAA ATT CGA TTG | 209 |
| CTT TCC AGC CTA CTT TTT TAT CAG T | |||
| hlB | GTG CAC TTA CTG ACA ATA GTG C | 309 | |
| GTT GAT GAG TAG CTA CCT TCA GT | |||
| 纤连蛋白结合蛋白 Fibronectin-binding protein | fnbA | GTG AAG TTT TAG AAG GTG GAAAGA TTA G | 634 |
| GCT CTT GTAAGA CCA TTT TTC TTC AC | |||
| fnbB | GTAACA GCT AAT GGT CGA ATT GAT ACT | 524 | |
| CAA GTT CGA TAG GAG TAC TAT GTT C | |||
| 黏附因子 Adhesion factor | clfA | ATT GGC GTG GCT TCA GTG CT | 292 |
| CGT TTC TTC CGT AGT TGC ATT TG | |||
| clfB | TGC AAG TGC AGA TTC CGA AAA AAA C | 194 | |
| CCG TCG GTT GAG GTG TTT CAT TTG | |||
| 胶原蛋白黏附素 Collagen adhesin | cnA | AAA GCG TTG CCT AGT GGA GAC | 228 |
| AGT GCC TTC CCA AAC CTT TT | |||
| 多重耐药基因 Multidrug-resistant gene | Cfr | TGAAGTATAAAGCAGGTTGGGAGTCA | 746 |
| ACCATATAATTGACCACAAGCAGC | |||
| 氨基糖苷类 Aminoglycoside | aac(6′)/aph(2′) | CCAAGAGCAATAAGGGCATA | 220 |
| CACTATCATAACCACTACCG | |||
| aph(3′)-Ⅲa | CACTATCATAACCACTACCG | 523 | |
| GCCGATGTGGATTGCGAAAA | |||
| ant(4′) | GCAAGGACCGACAACATTTC | 294 | |
| TGGCACAGATGGTCATAACC | |||
| ant(6′) | ACTGCTTAATCAATTGG | 597 | |
| GCCTTTCCGCCACCTCACCG | |||
| ant(3′) | TGATTTGCTGGTTACGGTGAC | 284 | |
| CGCTATGTTCTCTTGCTTTTG | |||
| 大环内酯类 Macrolide | ermA | TCAGGAAAAGGACATTTTACC | 423 |
| ATACTTTTTGTAGTCCTTCTT | |||
| ermB | TGGTATTCCAAATGCGTAATG | 745 | |
| CTGTGGTATGGCGGGTAAGT | |||
| ermC | CGTAACTGCCATTGAAATAGACC | 246 | |
| AATAATCTTTTAGCAAACCCGTA | |||
| 四环素类 Tetracycline | tetM | GTGGACAAAGGTACAACGAG | 406 |
| CGGTAAAGTTCGTCACACAC | |||
| tetO | AACTTAGGCATTCTGGCTCAC | 515 | |
| TCCCACTGTTCCATATCGTCA | |||
| 万古霉素 Vancomycin | vanA | GCTATTCAGCTGTACTC | 783 |
| CAGCGGCCATCATACGG | |||
| vanB | CATCGCCGTCCCCGAATTTCAAA | 297 | |
| GATGCGGAAGATACCGTGGCT | |||
| vanC | GGTATCAAGGAAACCTC | 822 | |
| CTTCCGCCATCATAGCT | |||
| β-内酰胺类 β-lactam | femA | TTACTACGCTGGTGGAACTTCA | 191 |
| CATCGGCATCATAGCCCTTT |
Table 2
Criteria for determining drug sensitivity"
抗菌药物 Antibacterial drug | 抑菌圈直径/mm Inhibition circle diameter | ||
| 敏感(S) Sensitivity | 中介(I) Intermediary | 耐药(R) Resistance | |
| 克林霉素 Clindamycin | ≥21 | 15~20 | ≤14 |
| 庆大霉素 Gentamicin | ≥15 | 13~14 | ≤12 |
| 四环素 Tetracycline | ≥19 | 15~18 | ≤14 |
| 青霉素G Penicillin G | ≥29 | — | ≤28 |
| 红霉素 Erythromycin | ≥23 | 14~22 | ≤13 |
| 诺氟沙星 Norfloxacin | ≥17 | 13~16 | ≤12 |
| 利福平 Rifampicin | ≥20 | 17~19 | ≤16 |
| 复方新诺明 Compound sulfamethoxazole | ≥17 | 13~16 | ≤12 |
| 氯霉素 Chloramphenicol | ≥18 | 13~17 | ≤12 |
| 头孢西丁 Cefoxitin | ≥22 | — | ≤21 |
Table 3
Staphylococcus aureus drug susceptibility results"
| 抗生素分类 Classification of antibiotics | 药物名称 Drug name | 敏感菌株比例/% Percentage of sensitivity (%)(n/n) | 中介菌株比例/% Percentage of intermediate (%)(n/n) | 耐药菌株比例/% Percentage of resistance(%)(n/n) |
| 林可霉素类 Lincomamides | 克林霉素 | 0.0(0/78) | 10.3(8/78) | 89.7(70/78) |
| 氨基糖苷类 Aminoglycosides | 庆大霉素 | 14.1(11/78) | 3.8(3/78) | 82.1(64/78) |
| 四环素类 Tetracyclines | 四环素 | 0.0(0/78) | 0.0(0/78) | 100.0(78/78) |
| β-内酰胺类 β-lactam class | 青霉素G | 1.3(1/78) | 0.0(0/78) | 98.7(77/78) |
| 头孢西丁 | 100.0(78/78) | 0.0(0/78) | 0.0(0/78) | |
| 大环内酯类 Macrolides | 红霉素 | 17.9(14/78) | 2.6(2/78) | 79.5(62/78) |
| 氟喹诺酮类 Fluoroquinolones | 诺氟沙星 | 2.6(2/78) | 1.3(1/78) | 96.2(75/78) |
| 安莎霉素类 Ansamycins | 利福平 | 97.4(76/78) | 0.0(0/78) | 2.6(2/78) |
| 叶酸途径拮抗剂 Folic acid antagonists | 复方新诺明 | 14.1(11/78) | 1.3(1/78) | 84.6(66/78) |
| 氯霉素类 Chloramphenicols | 氯霉素 | 30.8(24/78) | 23.1(18/78) | 46.2(36/78) |
Table 4
Results of multidrug resistance in Staphylococcus aureus"
| 耐药菌株号 Resistant strain number | 耐药重数 Multiple | 耐药菌株数 Number of resistance | 耐药菌株比例/% Percentage of resistant strains |
| LN-68 | 9 | 1 | 1.3 |
| LN-1,4,7,9,13,14,15,18,21,26,29,31,33,34,35,45,46,47,49,52,53,55,57,60,63,65,66,70,71,72 | 8 | 30 | 38.5 |
| LN-3,8,16,17,20,22,25,28,32,39,44,56,69,73,74,75,76 | 7 | 17 | 21.8 |
| LN-10,19,24,36,38,41,43,51,54,58,61,62,67 | 6 | 13 | 16.7 |
| LN-2,5,6,11,12,23,27,30,37,40,42,50,59,64,77,78 | 5 | 16 | 20.5 |
| LN-48 | 4 | 1 | 1.3 |
Fig. 7
Biofilm formation ability detection of some strains ODc=mean OD value of negative control+3s, OD570 nm >4 ODc for strong film-forming ability, 2 ODc < OD570 nm≤4 ODc for medium film-forming ability, 1 ODc < OD570 nm≤2 ODc for weak film-forming ability, OD570 nm≤1 ODc for no film-forming ability"
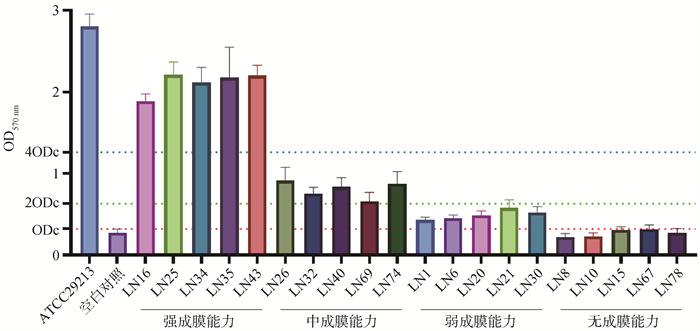
Table 5
Analysis results of drug resistance correlation data"
| 基因类别 Gene category | 基因名称 Gene name | 药物名称 Drug name | 敏感/% Sensitive | 中介/% Intermediate resistance | 耐药/% Resistant | χ2 | P |
| 氨基糖苷类 Aminoglycosides | aac(6′)/aph(2′) | 庆大霉素 | 5/11(45.45) | 3/3(100.00) | 64/64(100.00) | 39.591 | 0.000** |
| ant(4′) | 11/11(100.00) | 1/3(33.33) | 51/64(79.69) | 7.013 | 0.030* | ||
| 大环内酯类 Macrolides | ermB | 红霉素 | 1/14(7.14) | 0/2(0.00) | 38/62(61.29) | 15.447 | 0.000** |
| ermC | 1/14(7.14) | 1/2(50.00) | 40/62(64.52) | 15.14 | 0.001** | ||
| 多重耐药类 Multi drug resistance class | cfr | 氯霉素 | 0/24(0.00) | 0/18(0.00) | 1/36(2.78) | 1.182 | 0.554 |
| 复方新诺明 | 0/11(0.00) | 0/11(0.00) | 1/66(1.52) | 0.184 | 0.912 | ||
| 利福平 | 1/76(1.32) | 0/0(0.00) | 0/2(0.00) | 0.027 | 0.87 | ||
| 诺氟沙星 | 0/2(0.00) | 0/1(0.00) | 1/75(1.33) | 0.041 | 0.98 | ||
| 红霉素 | 0/14(0.00) | 1/2(50.00) | 0/62(0.00) | 38.494 | 0.000** | ||
| 庆大霉素 | 0/11(0.00) | 0/3(0.00) | 1/64(1.56) | 0.222 | 0.895 | ||
| 克林霉素 | 0/0(0.00) | 0/8(0.00) | 1/70(1.43) | 0.116 | 0.734 | ||
| 青霉素G | 1/1(100.00) | 0/0(0.00) | 0/77(0.00) | 78 | 0.000** |
Table 6
Results of correlation analysis between biofilm phenotypes and drug resistance phenotypes"
| 抗生素 Antibiotics | 分类 Classification | 生物被膜/% Biofilm | 总计 Total | χ2 | P | |||
| 无 None | 弱 Weak | 中 Moderate | 强 Strong | |||||
克林霉素 Clindamycin | 敏感Sensitivity | 0(0.00) | 0(0.00) | 0(0.00) | 1(4.00) | 1(1.28) | 4.343 | 0.227 |
| 中介Intermediary | 0(0.00) | 1(3.85) | 2(9.09) | 5(20.00) | 8(10.26) | |||
| 耐药Resistance | 5(100.00) | 25(96.15) | 20(90.91) | 20(80.00) | 70(89.74) | |||
| 总计Total | 5 | 26 | 22 | 25 | 78 | |||
庆大霉素 Gentamicin | 敏感Sensitivity | 1(20.00) | 4(15.38) | 5(22.73) | 1(4.00) | 11(14.10) | 6.967 | 0.324 |
| 中介Intermediary | 0(0.00) | 1(3.85) | 2(9.09) | 0(0.00) | 3(3.85) | |||
| 耐药Resistance | 4(80.00) | 21(80.77) | 15(68.18) | 24(96.00) | 64(82.05) | |||
| 总计Total | 5 | 26 | 22 | 25 | 78 | |||
青霉素G Penicillin G | 敏感Sensitivity | 0(0.00) | 0(0.00) | 0(0.00) | 1(4.00) | 1(1.28) | 2.148 | 0.542 |
| 中介Intermediary | 0(0.00) | 0(0.00) | 0(0.00) | 1(4.00) | 1(1.28) | |||
| 耐药Resistance | 5(100.00) | 26(100.00) | 22(100.00) | 24(96.00) | 77(98.72) | |||
| 总计Total | 5 | 26 | 22 | 25 | 78 | |||
红霉素 Erythromycin | 敏感Sensitivity | 0(0.00) | 1(3.85) | 2(9.09) | 11(44.00) | 14(17.95) | 23.086 | 0.001** |
| 中介Intermediary | 0(0.00) | 0(0.00) | 0(0.00) | 2(8.00) | 2(2.56) | |||
| 耐药Resistance | 5(100.00) | 25(96.15) | 20(90.91) | 12(48.00) | 62(79.49) | |||
| 总计Total | 5 | 26 | 22 | 25 | 78 | |||
诺氟沙星 Norfloxacin | 敏感Sensitivity | 0(0.00) | 0(0.00) | 1(4.55) | 1(4.00) | 2(2.56) | 3.546 | 0.738 |
| 中介Intermediary | 0(0.00) | 0(0.00) | 0(0.00) | 1(4.00) | 1(1.28) | |||
| 耐药Resistance | 5(100.00) | 26(100.00) | 21(95.45) | 23(92.00) | 75(96.15) | |||
| 总计Total | 5 | 26 | 22 | 25 | 78 | |||
利福平 Rifampicin | 敏感Sensitivity | 5(100.00) | 26(100.00) | 21(95.45) | 24(96.00) | 76(97.44) | 1.368 | 0.713 |
| 中介Intermediary | 0(0.00) | 0(0.00) | 0(0.00) | 1(4.00) | 1(1.28) | |||
| 耐药Resistance | 0(0.00) | 0(0.00) | 1(4.55) | 1(4.00) | 2(2.56) | |||
| 总计Total | 5 | 26 | 22 | 25 | 78 | |||
复方新诺明 Compound sulfamethoxazole | 敏感Sensitivity | 1(20.00) | 4(15.38) | 5(22.73) | 1(4.00) | 11(14.10) | 6.432 | 0.377 |
| 中介Intermediary | 0(0.00) | 0(0.00) | 1(4.55) | 0(0.00) | 1(1.28) | |||
| 耐药Resistance | 4(80.00) | 22(84.62) | 16(72.73) | 24(96.00) | 66(84.62) | |||
| 总计Total | 5 | 26 | 22 | 25 | 78 | |||
氯霉素 Chloramphenicol | 敏感Sensitivity | 1(20.00) | 6(23.08) | 8(36.36) | 9(36.00) | 24(30.77) | 26.915 | 0.000** |
| 中介Intermediary | 2(40.00) | 2(7.69) | 1(4.55) | 13(52.00) | 18(23.08) | |||
| 耐药Resistance | 2(40.00) | 18(69.23) | 13(59.09) | 3(12.00) | 36(46.15) | |||
| 总计Total | 5 | 26 | 22 | 25 | 78 | |||
| 1 |
MCNAMEEP T,SMYTHJ A.Bacterial chondronecrosis with osteomyelitis ('femoral head necrosis') of broiler chickens: a review[J].Avian Pathol,2000,29(4):253-270.
doi: 10.1080/03079450050118386 |
| 2 |
NAIRNM E,WATSONA R.Leg weakness of poultry--a clinical and pathological characterisation[J].Aust Vet J,1972,48(12):645-656.
doi: 10.1111/j.1751-0813.1972.tb09237.x |
| 3 |
WYERSM,CHERELY,PLASSIARTG.Late clinical expression of lameness related to associated osteomyelitis and tibial dyschondroplasia in male breeding turkeys[J].Avian Dis,1991,35(2):408-414.
doi: 10.2307/1591199 |
| 4 |
CHAOG,ZHANGX,ZHANGX,et al.Phenotypic and genotypic characterization of methicillin-resistant Staphylococcus aureus (MRSA) and methicillin-susceptible Staphylococcus aureus (MSSA) from different sources in China[J].Foodborne Pathog Dis,2013,10(3):214-221.
doi: 10.1089/fpd.2012.1205 |
| 5 |
DONGQ,WANGQ,ZHANGY,et al.Prevalence, antimicrobial resistance, and staphylococcal toxin genes of bla(TEM-1a)-producing Staphylococcus aureus isolated from animals in Chongqing, China[J].Vet Med Sci,2023,9(1):513-522.
doi: 10.1002/vms3.1028 |
| 6 | 洪捷,曹阳,谈忠鸣,等.江苏地区金黄色葡萄球菌耐药及分子分型研究[J].中国人兽共患病学报,2019,35(7):604-609. |
| HONGJ,CAOY,TANZ M,et al.Study on drug resistance and molecular typing of Staphylococcus aureus in Jiangsu region[J].Chinese Journal of Zoonoses,2019,35(7):604-609. | |
| 7 | 李新圃,王胜义,杨峰,等.金黄色葡萄球菌生物膜感染机制及药物作用研究进展[J].动物医学进展,2020,41(4):99-103. |
| LIX P,WANGS Y,YANGF,et al.Research progress on the infection mechanisms of Staphylococcus aureus biofilms and drug effects[J].Progress in Veterinary Medicine,2020,41(4):99-103. | |
| 8 |
LOWDERB V,GUINANEC M,BEN ZAKOURN L,et al.Recent human-to-poultry host jump, adaptation, and pandemic spread of Staphylococcus aureus[J].Proc Natl Acad Sci U S A,2009,106(46):19545-19550.
doi: 10.1073/pnas.0909285106 |
| 9 |
SZAFRANIECG M,SZELESZCZUKP,DOLKAB.Review on skeletal disorders caused by Staphylococcus spp. in poultry[J].Vet Q,2022,42(1):21-40.
doi: 10.1080/01652176.2022.2033880 |
| 10 | MONTANAROL,RAVAIOLIS,RUPPITSCHW,et al.Molecular Characterization of a prevalent ribocluster of methicillin-sensitive Staphylococcus aureus from orthopedic implant infections. Correspondence with MLST CC30[J].Front Cell Infect Microbiol,2016,6,8. |
| 11 |
KÄPPELIN,MORACHM,CORTIS,et al.Staphylococcus aureus related to bovine mastitis in Switzerland: Clonal diversity, virulence gene profiles, and antimicrobial resistance of isolates collected throughout 2017[J].J Dairy Sci,2019,102(4):3274-3281.
doi: 10.3168/jds.2018-15317 |
| 12 | 杨慧君,王艺晖,李晓娜,等.宁夏地区牛源金黄色葡萄球菌毒力基因和大环内酯类抗生素耐药基因的检测及诱导耐药[J].中国兽医学报,2018,38(5):962-967, 985. |
| YANGH J,WANGY H,LIX N,et al.Detection of virulence genes and macrolide resistance genes in bovine Staphylococcus aureus in Ningxia region and induced drug resistance[J].Chinese Journal of Veterinary Science,2018,38(5):962-967, 985. | |
| 13 |
MANNINGS D,KIM,MARRSC F,et al.The frequency of genes encoding three putative group B streptococcal virulence factors among invasive and colonizing isolates[J].BMC Infect Dis,2006,6,116.
doi: 10.1186/1471-2334-6-116 |
| 14 |
CHUANCHUENR,PADUNGTODP.Antimicrobial resistance genes in Salmonella enterica isolates from poultry and swine in Thailand[J].J Vet Med Sci,2009,71(10):1349-1355.
doi: 10.1292/jvms.001349 |
| 15 |
POETAP,COSTAD,SÁENZY,et al.Characterization of antibiotic resistance genes and virulence factors in faecal enterococci of wild animals in Portugal[J].J Vet Med B Infect Dis Vet Public Health,2005,52(9):396-402.
doi: 10.1111/j.1439-0450.2005.00881.x |
| 16 |
KIMD H,CHUNGY S,PARKY K,et al.Antimicrobial resistance and virulence profiles of Enterococcus spp. isolated from horses in korea[J].Comp Immunol Microbiol Infect Dis,2016,48,6-13.
doi: 10.1016/j.cimid.2016.07.001 |
| 17 | 谢常红,刘桂兰,王颖,等.鸡源金黄色葡萄球菌、大肠杆菌、奇异变形杆菌的分离鉴定及生物学特性研究[J].中国畜牧兽医,2024(12):5549-5559. |
| XIEC H,LIUG L,WANGY,et al.Isolation, identification and biological characteristics of Staphylococcus aureus, Escherichia coli and Proteus mirabilis from poultry[J].China Animal Husbandry & Veterinary Medicine,2024(12):5549-5559. | |
| 18 |
PIMENTEL DE ARAUJOF,PIROLOM,MONACOM,et al.Virulence determinants in Staphylococcus aureus clones causing osteomyelitis in Italy[J].Front Microbiol,2022,13,846167.
doi: 10.3389/fmicb.2022.846167 |
| 19 | ZAREI KOOSHAR,MAHMOODZADEH HOSSEINIH,MEHDIZADEH AGHDAME,et al.Distribution of tsst-1 and mecA genes in Staphylococcus aureus isolated from clinical specimens[J].Jundishapur J Microbiol,2016,9(3):e29057. |
| 20 |
ZENGM,GUOZ,SHENS,et al.Value of serum procalcitonin and interleukin-6 in patients with bullous impetigo and staphylococcal scalded skin syndrome[J].J Dermatol,2014,41(11):1028-1029.
doi: 10.1111/1346-8138.12638 |
| 21 | 陶欣月,马曦.饲用贝莱斯芽孢杆菌的分离及竞争性抑制金黄色葡萄球菌粘附的研究[J].中国畜牧杂志,2022,58(12):240-246. |
| TAOX Y,MAX.Isolation of feed-grade Bacillus velezensis and its competitive inhibition against Staphylococcus aureus adhesion[J].Chinese Journal of Animal Science,2022,58(12):240-246. | |
| 22 |
LOZANOC,GHARSAH,BEN SLAMAK,et al. Staphylococcus aureus in animals and food: methicillin resistance, prevalence and population structure. A Review in the African continent[J].Microorganisms,2016,4(1):12.
doi: 10.3390/microorganisms4010012 |
| 23 | ALONZO F, 3rd, TORRES V J. The bicomponent pore-forming leucocidins of Staphylococcus aureus[J]. Microbiol Mol Biol Rev, 2014, 78(2): 199-230. |
| 24 |
NHANT X,LECLERCQR,CATTOIRV.Prevalence of toxin genes in consecutive clinical isolates of Staphylococcus aureus and clinical impact[J].Eur J Clin Microbiol Infect Dis,2011,30(6):719-725.
doi: 10.1007/s10096-010-1143-4 |
| 25 |
LEEA S,DE LENCASTREH,GARAUJ,et al.Methicillin-resistant Staphylococcus aureus[J].Nat Rev Dis Primers,2018,4,18033.
doi: 10.1038/nrdp.2018.33 |
| 26 | 朱德妹,汪复,胡付品,等.2010年中国CHINET细菌耐药性监测[J].中国感染与化疗杂志,2011,11(5):321-329. |
| ZHUD M,WANGF,HUF P,et al.Surveillance of bacterial resistance in China CHINET in 2010[J].Chinese Journal of Infection and Chemotherapy,2011,11(5):321-329. | |
| 27 |
XUZ,CHENX,TANW,et al.Prevalence and antimicrobial resistance of Salmonella and Staphylococcus aureus in fattening pigs in Hubei Province, China[J].Microb Drug Resist,2021,27(11):1594-1602.
doi: 10.1089/mdr.2020.0585 |
| 28 |
LIUY,ZHENGX,XUL,et al.Prevalence, antimicrobial resistance, and molecular characterization of Staphylococcus aureus isolated from animals, meats, and market environments in Xinjiang, China[J].Foodborne Pathog Dis,2021,18(10):718-726.
doi: 10.1089/fpd.2020.2863 |
| 29 |
PUW,SUY,LIJ,et al.High incidence of oxacillin-susceptible mecA-positive Staphylococcus aureus (OS-MRSA) associated with bovine mastitis in China[J].PLoS One,2014,9(2):e88134.
doi: 10.1371/journal.pone.0088134 |
| 30 |
崔恒洁,覃金珑,朱志豪,等.金黄色葡萄球菌对苯扎溴铵敏感性与生物被膜形成能力相关性分析[J].畜牧兽医学报,2024,55(8):3669-3677.
doi: 10.11843/j.issn.0366-6964.2024.08.038 |
|
CUIH J,QINJ L,ZHUZ H,et al.Correlation analysis of benzalkonium bromide sensitivity and biofilm formation ability in Staphylococcus aureus[J].Acta Veterinaria et Zootechnica Sinica,2024,55(8):3669-3677.
doi: 10.11843/j.issn.0366-6964.2024.08.038 |
|
| 31 |
HALL-STOODLEYL,COSTERTONJ W,STOODLEYP.Bacterial biofilms: from the natural environment to infectious diseases[J].Nat Rev Microbiol,2004,2(2):95-108.
doi: 10.1038/nrmicro821 |
| 32 |
CAMPOCCIAD,MIRZAEIR,MONTANAROL,et al.Hijacking of immune defences by biofilms: a multifront strategy[J].Biofouling,2019,35(10):1055-1074.
doi: 10.1080/08927014.2019.1689964 |
| 33 |
ARCHERN K,MAZAITISM J,COSTERTONJ W,et al.Staphylococcus aureus biofilms: properties, regulation, and roles in human disease[J].Virulence,2011,2(5):445-459.
doi: 10.4161/viru.2.5.17724 |
| 34 |
JIANGX,HED,GAOL,et al.Synergistic pathogenicity of avian orthoreovirus and Staphylococcus aureus on SPF chickens[J].Poult Sci,2023,102(10):102996.
doi: 10.1016/j.psj.2023.102996 |
| 35 | 高玲. 鸡呼肠孤病毒和金黄色葡萄球菌对SPF鸡的协同致病作用研究[D]. 泰安: 山东农业大学, 2022. |
| GAO L. Synergistic pathogenic effects of avian reovirus and Staphylococcus aureus in SPF chickens[D]. Taian: Shandong Agricultural University, 2022. (in Chinese) |
| [1] | YANG Shubo, YUAN Qingxin, CHEN Qibai, WANG Pei, GAO Dongyang, LI He, SONG Jun. Preparation and Intracellular Antibacterial Activity of Liposomes Encapsulating Staphylococcus aureus Bacteriophage [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(9): 4638-4645. |
| [2] | SHI Wenjian, XU Lei, ZHANG Ze, YANG Rui, XIN Lingxiang, WANG Nan, CHEN Xiang, XIN Ting. Screening and Identification of Biofilm-related Genes Based on the Random Mutant Library of Mycobacterium tuberculosis Variant bovis C68001 [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(8): 3992-4006. |
| [3] | XIANG Lingxian, JI Qianyu, SHAN Xinxin, LI Lin. Advances in the Study of Drug Resistance and Pathogenicity of Bacterial Two-component Systems [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(7): 3116-3128. |
| [4] | KONG Xianghe, SU Xinyu, ZHONG Yiming, LUZHAO Zixuan, LIAO Xianmao, ZHANG Zhe, ZHANG Xu, GAO Mengmeng, ZHOU Yulong, FAN Huqing. To Investigate the Prevalence, Virulence Genes and Drug Resistance of Pathogenic Escherichia coli Isolated from Calves with Diarrhea in Heilongjiang Province [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(7): 3390-3398. |
| [5] | LIU Yuze, YU Zhuoya, GONG Zhiguo, REN Peipei, ZHAO Jiamin, MAO Wei, ZHANG Shuangyi, FENG Shuang. The Impact of Lipoprotein on the Secretion of Inflammatory Mediators and the Synthesis of Prostaglandin E2 in Bovine Bone Marrow-derived Macrophages Infected with Staphylococcus aureus [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(7): 3474-3483. |
| [6] | ZHAO Enhao, SHI Hongmei, GESANG Zhuoma, SUOLANG Sizhu, GONG Ga. Genetic Evolution, Virulence Genes, and Drug Resistance Analysis of Klebsiella pneumoniae from Yak in Gansu Province [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(6): 2893-2905. |
| [7] | ZHANG Manqi, ZHAO Bingyu, WEN Ruru, ZHANG Jingwen, SUN Mengran, ZHAN Leyang, GOU Jingxuan, SONG Xiangjun. Prokaryotic Expression of the T6SS Effector Protein Tse1 and Its inhibitory Effect on Staphylococcus aureus [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(6): 2917-2926. |
| [8] | JI Xing, LI Jun, WANG Ran, HE Tao. Research Progress on Virulence Regulation and Antivirulence Drugs of Staphylococcus aureus [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(4): 1594-1607. |
| [9] | LIAO Yiwen, YE Jingfen, WU Shaobi, CHEN Shixiong, YANG Wan, LUO Xue, YANG Qi. Development of Ring-mediated Isothermal Amplification Technology and Its Application to the Detection of Drug Resistance Genes [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(4): 1621-1631. |
| [10] | LI Anben, FU Nana, LUO Xiaoping, LI Junyan, LIU Yang. Progress in the Study of Drug Resistance and Its Reversal in Haemonchus contortus [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(2): 523-533. |
| [11] | LI Zhenya, LIU Jie, LI Yun, WANG Fei, KONG Yuanyuan, LI Yong, JIA Rongling. Biological Characteristics and Comparative Genomic Analysis of Virulent and Attenuated Strains of Mycoplasma hyopneumoniae [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(2): 851-859. |
| [12] | DU Qingjie, WU Liping, ZHANG Fan, DAI Pengxiu, FENG Xiancheng, ZHANG Xinke. Difference Analysis of Oral Flora in Dogs with Periodontitis and Drug Resistance of Oral Porphyromonas [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(2): 934-942. |
| [13] | BAI Yixin, ZHANG Mengfei, Dinala·Yeerboli , WANG Lei, JIN Wanjing, HUANG Ying, XIE Jinxin, ZHENG Xiaofeng, WANG Meiling, WANG Chuanfeng, SU Zhanqiang, ZHANG Wei, TONG Panpan. Analysis of Virulence Genes, Drug Resistance and Biofilm of Diarrheagenic Escherichia coli from Young Livestock in Xinjiang [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(10): 5125-5136. |
| [14] | YU Bingyu, YE Yitong, CHEN Yixing, ZHANG Meilin, YANG Zhican, CHEN Jidang, ZHU Wanjun, ZHANG Jipei. Antimicrobial Resistance Analysis of Escherichia coli from Goose in Guangdong [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(10): 5202-5211. |
| [15] | ZHANG Lei, CHEN Liang, FENG Wanyu, LAN Shijie, MIAO Yan, TIAN Qiufeng, BAI Changsheng, ZHANG Bei, DONG Jiaqiang, JIANG Botao, WANG Hongbao, SHI Tongrui, HUANG Xuankai. The Role of Biofilms in the Pathogenesis of Animal Bacterial Infections [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(1): 107-114. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||