





Acta Veterinaria et Zootechnica Sinica ›› 2025, Vol. 56 ›› Issue (7): 3226-3243.doi: 10.11843/j.issn.0366-6964.2025.07.017
• Animal Genetics and Breeding • Previous Articles Next Articles
BAI Yuanyuan1( ), CAI Wenyi1, XING Jiayi1, JIANG Yuting1, MA Zhiwei1, JI Wenhui1,2,*(
), CAI Wenyi1, XING Jiayi1, JIANG Yuting1, MA Zhiwei1, JI Wenhui1,2,*( ), LAN Daoliang1,2,*(
), LAN Daoliang1,2,*( )
)
Received:2024-10-28
Online:2025-07-23
Published:2025-07-25
Contact:
JI Wenhui, LAN Daoliang
E-mail:2934245090@qq.com;somebody528@163.com;landaoliang@163.com
CLC Number:
BAI Yuanyuan, CAI Wenyi, XING Jiayi, JIANG Yuting, MA Zhiwei, JI Wenhui, LAN Daoliang. Comparative Transcriptome Mapping of the Lungs of Yak, Dzho, and Cattle[J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(7): 3226-3243.
Table 1
Databases and version information"
| 数据库 Database | 网页链接 Web links | 版本信息 Version information |
| mRNA数据库 mRNA database | ARS_UCD1.2_NCBI | |
| KEGG数据库 KEGG database | NA | |
| GO数据库GO database | NA | |
| 基因组数据库 Genome database | ARS_UCD1.2_NCBI | |
| 基因组注释文件 Genome annotation files | ARS_UCD1.2_NCBI |
Table 2
Primer sequences used for real-time PCR"
| 引物名称 Primer name | 引物序列(5′→3′) Primer sequence | 产物长度/bp Size |
| GAPDH | F: AAGGTCGGAGTGAACGGATT;R: TTGATGACGAGCTTCCCGTT | 197 |
| ALAS2 | R: TTGATGACGAGCTTCCCGTT;F: TACGTTCCTATGCTGCTGGC | 140 |
| ALDH3A1 | F: CATCCTTACGGACGTGGACC;R: TCACCTTGTCGTTCGGTGAG | 164 |
| SYT13 | F: GCACCCCAAGAAGGGAGTGT;R: GGACGGGCTCTGTGGACTT | 114 |
| DEFB7 | F: CTCTTCCTGGTCCTGTCTG;R: GTGCCAATCTGTCTCCTGT | 146 |
| CHRNA7 | F: TTGGGTCCTGGTCTTATGG;R: TCTGGGTAGGGTTCTTTGC | 160 |
Fig. 2
Analysis of gene expression levels A. Box plot of gene expression level: blue for cattle, red for dzho, green for yak, horizontal axis is the sample name, vertical axis is log10 (FPKM+1), and the box plot of each region corresponds to 5 statistics (top to bottom, maximum, 3/4th quantile, median, 1/4th quartile and minimum). B. PCA diagram, the closer the sample clustering distance or PCA distance, the more similar the samples are. The horizontal and vertical coordinates PC1 and PC2 represent different principal components, and the percentages indicate the confidence with which the samples can be distinguished"
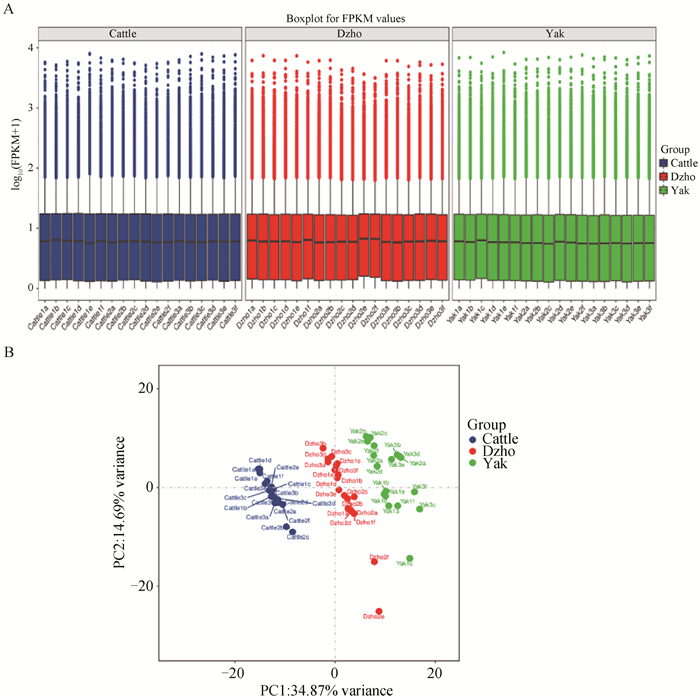
Fig. 4
GO and KEGG-pathway analysis of differentially expressed genes between yak and cattle A. The horizontal axis represents the name of GO terms, and the vertical axis represents -log10(P). B. The horizontal axis is the Enrichment Score, indicating the degree of enrichment, the larger the value, the higher the enrichment; Large bubbles indicate entries that contain more differentially expressed protein-coding genes; The bubble colors change from purple to blue to green to red, with closer proximity to red indicating smaller enrichment P-values and greater significance"
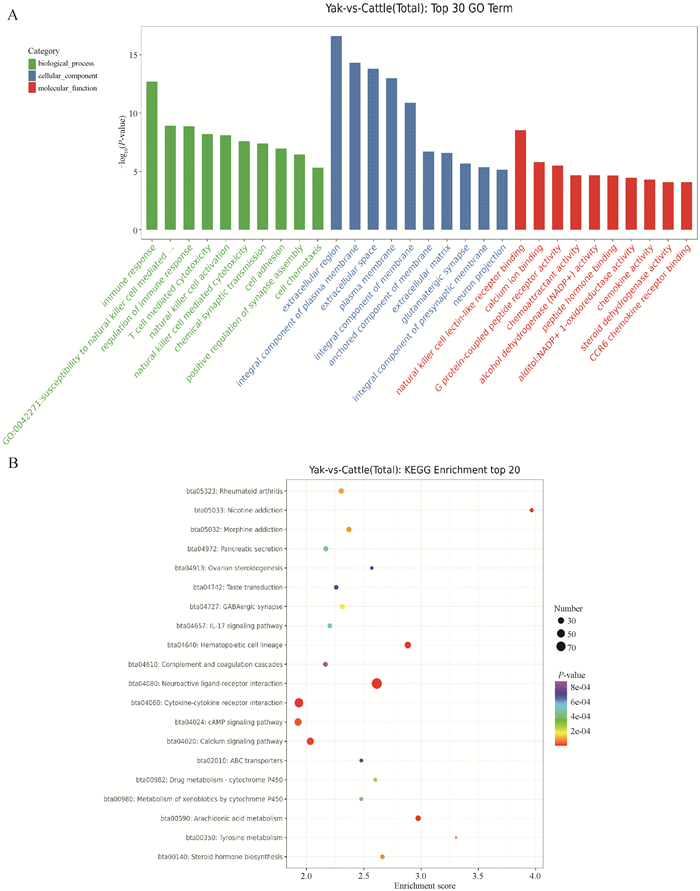
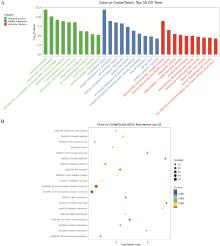
Fig. 5
GO and KEGG-pathway analysis of differentially expressed genes between dzho and cattle A. The horizontal axis represents the name of GO terms, and the vertical axis represents -log10(P). B. The horizontal axis is the Enrichment Score, indicating the degree of enrichment, the larger the value, the higher the enrichment; Large bubbles indicate entries that contain more differentially expressed protein-coding genes; The bubble colors change from purple to blue to green to red, with closer proximity to red indicating smaller enrichment P-values and greater significance"
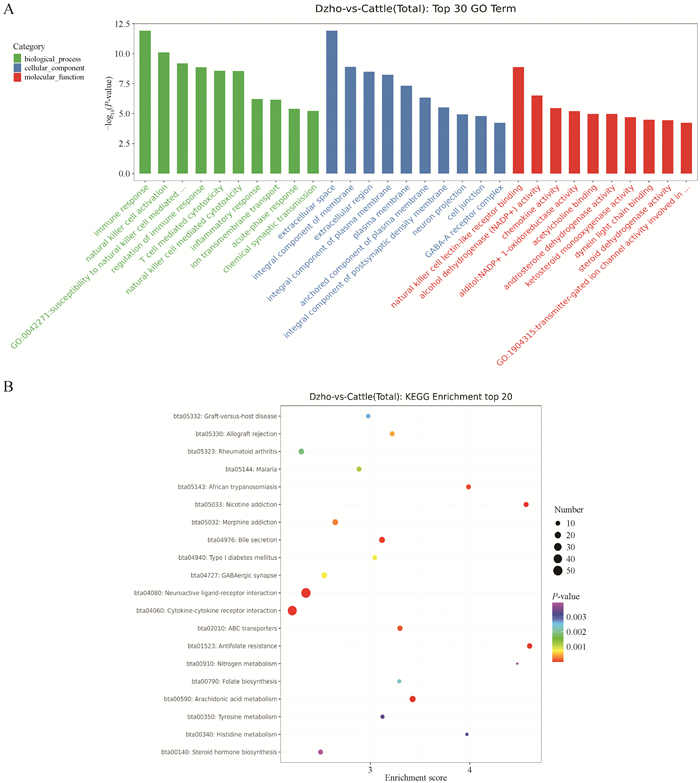
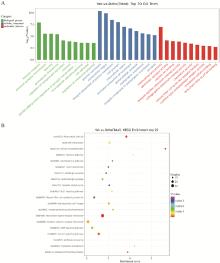
Fig. 6
GO and KEGG-pathway analysis of differentially expressed genes between yak and dzho A. The horizontal axis represents the name of GO terms, and the vertical axis represents -log10(P). B. The horizontal axis is the Enrichment Score, indicating the degree of enrichment, the larger the value, the higher the enrichment; Large bubbles indicate entries that contain more differentially expressed protein-coding genes; The bubble colors change from purple to blue to green to red, with closer proximity to red indicating smaller enrichment P-values and greater significance"
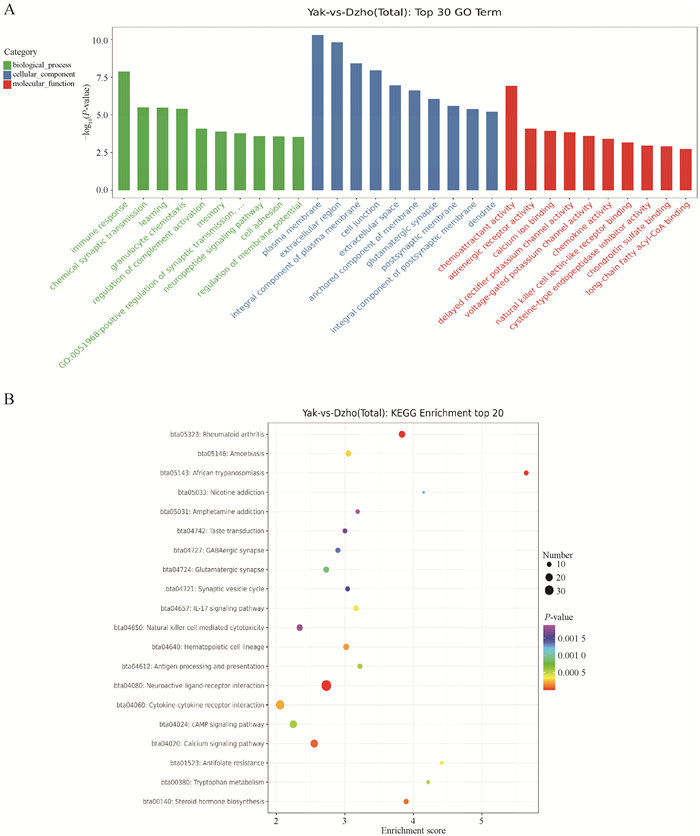

Fig. 8
GO and KEGG enrichment analysis of common differentially expressed genes A.The top 30 terms in the GO enrichment analysis: the horizontal axis is the -log10(P) and the vertical axis is the GO terms name.B.KEGG pathway enriched top 20 bubble map: the larger the bubble, the larger the number of genes encoding the differential proteins; the color of the bubble changes of violet-blue-green-red, and the smaller the enrichment P-value value, the greater the significance"
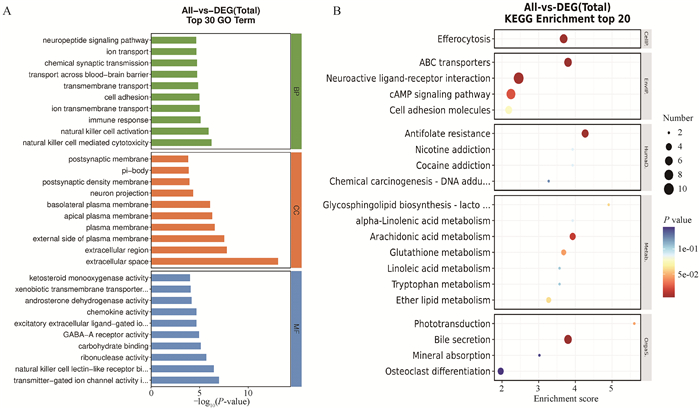

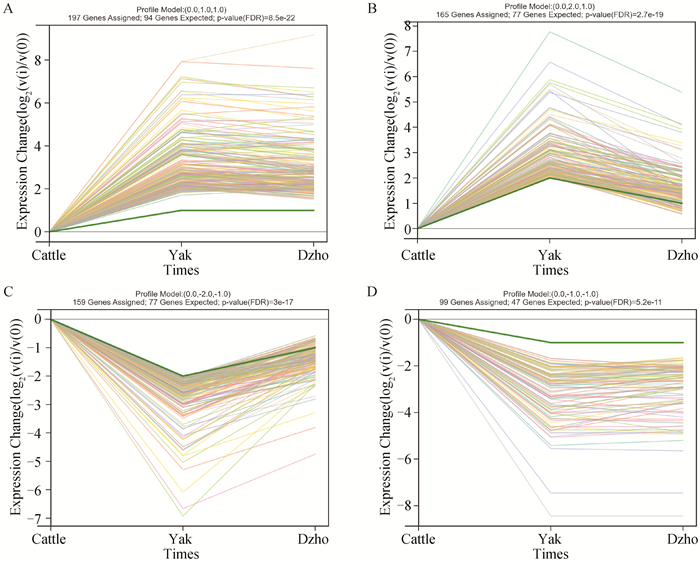
Fig. 9
Trend chart of significant modules In the module trend chart, the y-axis is the difference between each time node and the first time node, and the x-axis is the name of the time node, where the green thick line represents the trend of the module.In A, B figures, genes expressed in increasing amounts in the order of Cattle-Dzho-Yak; In C, D figures, genes are expressed in decreasing amounts in the order of Cattle-Dzho-Yak"
| 1 | PATRICIAN A , ANHOLM J D , AINSLIE P N . A narrative review of periodic breathing during sleep at high altitude: From acclimatizing lowlanders to adapted highlanders[J]. J Physiol, 2024, 602 (21): 5435- 5448. |
| 2 | LI Y , FRANDSEN K M , GUO W , et al. Impact of altitude on the dosage of indoor particulates entering an individual 's small airways[J]. J Hazard Mater, 2024, 468, 133856. |
| 3 | BÄRTSCH P , GIBBS J S . Effect of altitude on the heart and the lungs[J]. Circulation, 2007, 116 (19): 2191- 2202. |
| 4 | CHU M , GAO H , ESPARZA P , et al. Chronic developmental hypoxia alters rat lung immune cell transcriptomes during allergic airway inflammation[J]. Physiol Rep, 2023, 11 (3): e15600. |
| 5 | LI H , PEI W , WANG Y , et al. Mechanism of action of the plateau-adapted gene PPARA in COPD[J]. Front Biosci (Landmark Ed), 2024, 29 (2): 68. |
| 6 | SHARMA S , KOSHY R , KUMAR R , et al. Hypobaric hypoxia drives selection of altitude-associated adaptative alleles in the Himalayan population[J]. Sci Total Environ, 2024, 913, 169605. |
| 7 | YAN F , WANG Y , WEI M , et al. Exploring the role of the CapG gene in hypoxia adaptation in Tibetan pigs[J]. Front Genet, 2024, 15, 1339683. |
| 8 | GE Q , GUO Y , ZHENG W , et al. Molecular mechanisms detected in yak lung tissue via transcriptome-wide analysis provide insights into adaptation to high altitudes[J]. Sci Rep, 2021, 11 (1): 7786. |
| 9 | 何俊峰, 贺延玉, 崔燕. 牦牛心脏肺脏对高海拔低氧的适应性结构[C]. 郑州: 中国畜牧兽医学会动物解剖及组织胚胎学分会第十九次学术研讨会, 2016: 15. |
| HE J F, HE Y Y, CUI Y. Adaptive structure of yak heart and lungs to high altitude and low oxygen[C]. Zhengzhou: The 19th Academic Symposium of the Animal Anatomy and Tissue Embryology Branch of the Chinese Association of Animal Husbandry and Veterinary Medicine, 2016: 15. (in Chinese) | |
| 10 | WANG B , HE J , CUI Y , et al. The HIF-1α/EGF/EGFR signaling pathway facilitates the proliferation of yak alveolar type Ⅱ epithelial cells in hypoxic conditions[J]. Int J Mol Sci, 2024, 25 (3): 1442. |
| 11 | XIE X , WEI Y , CUI Y , et al. Transcriptomics reveals age-related changes in ion transport-related factors in yak lungs[J]. Front Vet Sci, 2024, 11, 1374794. |
| 12 | HE J , WEI Y , CUI Y , et al. Distribution and expression of pulmonary ionocyte-related factors CFTR, ATP6V0D2, and ATP6V1C2 in the lungs of yaks at different ages[J]. Genes (Basel), 2023, 14 (3): 597. |
| 13 | XIN J W , CHAI Z X , JIANG H , et al. Genome-wide comparison of DNA methylation patterns between yak and three cattle strains and their potential association with mRNA transcription[J]. J Exp Zool B Mol Dev Evol, 2023, 340 (4): 316- 328. |
| 14 | 杨德彬, 张燕, 金天虎, 等. 犏牛主要经济性状的研究进展[J]. 四川畜牧兽医, 2015, 42 (11): 36- 38. |
| YANG D B , ZHANG Y , JIN T H , et al. Research progress on the main economic traits of Pian cattle[J]. Sichuan Animal Husbandry and Veterinary Medicine, 2015, 42 (11): 36- 38. | |
| 15 | SLINGO M E . Oxygen-sensing pathways and the pulmonary circulation[J]. J Physiol, 2024, 602 (21): 5619- 5629. |
| 16 | 刘慧颖, 王浩, 李慧颖, 等. ABCA3参与的磷脂代谢在其相关肺疾病中的研究进展[J]. 临床肺科杂志, 2023, 28 (10): 1577- 1581. |
| LIU H Y , WANG H , LI H Y , et al. Research progress on ABCA3-mediated phospholipid metabolism in related pulmonary diseases[J]. Journal of Clinical Pulmonology, 2023, 28 (10): 1577- 1581. | |
| 17 | AILI A , WANG Y , SHANG Y , et al. LPG 18:0 is a general biomarker of asthma and inhibits the differentiation and function of regulatory T-cells[J]. Eur Respir J, 2024, 64 (6): 2301752. |
| 18 | LI N , SU S , XIE X , et al. Tsantan Sumtang, a traditional Tibetan medicine, protects pulmonary vascular endothelial function of hypoxia-induced pulmonary hypertension rats through AKT/eNOS signaling pathway[J]. J Ethnopharmacol, 2024, 320, 117436. |
| 19 | 王强, 陈静, 赵玉婷, 等. 探索α-亚麻酸的健康奥秘: 强化膳食, 延长生命质量[J]. 营养学报, 2023, 45 (4): 331- 336. |
| WANG Q , CHEN J , ZHAO Y T , et al. Exploring the health benefits of α-linolenic acid: Enhancing diet and prolonging life quality[J]. Acta Nutrimenta Sinica, 2023, 45 (4): 331- 336. | |
| 20 | ROHRS E C , BASSI T G , FERNANDEZ K C , et al. Diaphragm neurostimulation during mechanical ventilation reduces atelectasis and transpulmonary plateau pressure, preserving lung homogeneity and PaO2/FIO2[J]. J Appl Physiol, 2021, 131 (1): 290- 301. |
| 21 | PHILLIPS J D . Heme biosynthesis and the porphyrias[J]. Mol Genet Metab, 2019, 128 (3): 164- 177. |
| 22 | BELOT A , PUY H , HAMZA I , et al. Update on heme biosynthesis, tissue-specific regulation, heme transport, relation to iron metabolism and cellular energy[J]. Liver Int, 2024, 44 (9): 2235- 2250. |
| 23 | DUCAMP S , SENDAMARAI A K , CAMPAGNA D R , et al. Murine models of erythroid 5ALA synthesis disorders and their conditional synthetic lethal dependency on pyridoxine[J]. Blood, 2024, 144 (13): 1418- 1432. |
| 24 | DING Y , YANG K , LIU X , et al. A novel ALAS2 mutation causes congenital sideroblastic anemia[J]. Mediterr J Hematol Infect Dis, 2023, 15 (1): e2023062. |
| 25 | KHECHADURI A , BAYEVA M , CHANG H C , et al. Heme levels are increased in human failing hearts[J]. J Am Coll Cardiol, 2013, 61 (18): 1884- 1893. |
| 26 | HOFER T , WENGER R H , KRAMER M F , et al. Hypoxic upregulation of erythroid 5-aminolevulinate synthase[J]. Blood, 2003, 101 (1): 348- 350. |
| 27 | LORIA F , COX H D , VOSS S C , et al. The use of RNAbased 5'-aminolevulinate synthase 2 biomarkers in dried blood spots to detect recombinant human erythropoietin microdoses[J]. Drug Test Anal, 2022, 14 (5): 826- 832. |
| 28 | KHALIFA O , AL-AKL N S , ARREDOUANI A . Differential expression of cardiometabolic and inflammation markers and signaling pathways between overweight/obese Qatari adults with high and low plasma salivary α-amylase activity[J]. Front Endocrinol (Lausanne), 2024, 15, 1421358. |
| 29 | CHEN Y , YAN H , YAN L , et al. Hypoxia-induced ALDH3A1 promotes the proliferation of non-small-cell lung cancer by regulating energy metabolism reprogramming[J]. Cell Death Dis, 2023, 14 (9): 617. |
| 30 | ZHOU X , GUO Z , LIU S , et al. Transcriptomics and molecular docking reveal the potential mechanism of lycorine against pancreatic cancer[J]. Phytomedicine, 2024, 122, 155128. |
| 31 | LOU Y X , SHI E D , YANG R , et al. Exploring the mechanisms of glycolytic genes involvement in pulmonary arterial hypertension through integrative bioinformatics analysis[J]. J Cell Mol Med, 2024, 28 (11): e18447. |
| 32 | CHEN M , MAINARDI S , LIEFTINK C , et al. Targeting of vulnerabilities of drug-tolerant persisters identified through functional genetics delays tumor relapse[J]. Cell Rep Med, 2024, 5 (3): 101471. |
| 33 | WANG B , HE Y , WANG B , et al. ALDH3A1 overexpression in OSCC inhibits inflammation via phospho-Ser727 at STAT3 in tumor-associated macrophages[J]. Oral Dis, 2023, 29 (4): 1513- 1524. |
| 34 | SINGHRAO S K , CONSOLI C . Identifying Alzheimer 's disease genes in apolipoprotein E-/- mice brains with confirmed Porphyromonas gingivalis entry[J]. J Alzheimers Dis Rep, 2025, 9, 25424823251332874. |
| 35 | GODOY J A , LINDSAY C B , QUINTANILLA R A , et al. Quercetin exerts differential neuroprotective effects against H2O2 and Aβ aggregates in hippocampal neurons: the role of mitochondria[J]. Mol Neurobiol, 2017, 54 (9): 7116- 7128. |
| 36 | ZHANG Y D , ZHONG R , LIU J Q , et al. Role of synaptotagmin 13 (SYT13) in promoting breast cancer and signaling pathways[J]. Clin Transl Oncol, 2023, 25 (6): 1629- 1640. |
| 37 | 赵子瑜, 刘启玲, 姚金余, 等. 低氧导致小鼠注意缺陷多动障碍样行为及其突触损伤机制[J]. 空军军医大学学报, 2023, 44 (3): 229- 234. |
| ZHAO Z Y , LIU Q L , YAO J Y , et al. Attention deficit hyperactivity disorder-like behavior caused by hypoxia and its synaptic injury mechanism in mice[J]. Journal of Air Force Military Medical University, 2023, 44 (3): 229- 234. | |
| 38 | NIZZARDO M , TAIANA M , RIZZO F , et al. Synaptotagmin 13 is neuroprotective across motor neuron diseases[J]. Acta Neuropathol, 2020, 139 (5): 837- 853. |
| 39 | XU H , ZHANG L B , LUO Y Y , et al. Synaptotagmins family affect glucose transport in retinal pigment epithelial cells through their ubiquitination-mediated degradation and glucose transporter-1 regulation[J]. World J Diabetes, 2024, 15 (5): 958- 976. |
| 40 | MERRIMAN K E , POWELL J L , SANTOS J E P , et al. Intramammary 25-hydroxyvitamin D3 treatment modulates innate immune responses to endotoxin-induced mastitis[J]. J Dairy Sci, 2018, 101 (8): 7593- 7607. |
| 41 | DU J , RUI F , HAO Z , et al. Transcription factor E2F1 regulates the expression of ADRB2[J]. Int J Anal Chem, 2023, 2023, 8210685. |
| 42 | GAO X , WANG S , WANG Y F , et al. Long read genome assemblies complemented by single cell RNA-sequencing reveal genetic and cellular mechanisms underlying the adaptive evolution of yak[J]. Nat Commun, 2022, 13 (1): 4887. |
| 43 | AN Z F , ZHAO K , WEI L N , et al. p53 gene cloning and response to hypoxia in theplateau zokor, Myospalax baileyi[J]. Anim Biol, 2018, 68 (3): 289- 308. |
| 44 | 刘芳, 乌仁图雅, 马兰, 等. 高海拔低氧适应物种藏羚羊低氧诱导因子1Α的基因克隆与表达[J]. 生理学报, 2011, 63 (6): 565- 573. |
| LIU F , WUREN T , MA L , et al. , Genetic cloning and expression of hypoxia inducible factor 1 alpha in high altitude hypoxic adaptation species Tibetan antelope (Pantholops hodgsonii)[J]. Sheng Li Xue Bao, 2011, 63 (6): 565- 573. | |
| 45 | WANG H , LIU D , SONG P , et al. , Exposure to hypoxia causes stress erythropoiesis and downregulates immune response genes in spleen of mice[J]. BMC Genomics, 2021, 22 (1): 413. |
| 46 | HE X , SONG S , AYON R J , et al. Hypoxia selectively upregulates cation channels and increases cytosolic[Ca2+] in pulmonary, but not coronary, arterial smooth muscle cells[J]. Am J Physiol Cell Physiol, 2018, 314 (4): C504- C517. |
| 47 | PENG G , RAN P , LU W , et al. Acute hypoxia activates store-operated Ca(2+) entry and increases intracellular Ca(2+) concentration in rat distal pulmonary venous smooth muscle cells[J]. J Thorac Dis, 2013, 5 (5): 605- 612. |
| 48 | 姚敏. PLIγ重组表达载体构建及其抗LPS诱导的THP-1细胞炎症作用[D]. 南昌: 南昌大学, 2014. |
| YAO M. Construction of recombinant PLIγ expression vector and its anti-inflammatory effect on LPS-induced THP-1 cells[D]. Nanchang: Nanchang University, 2014. (in Chinese) |
| [1] | ZHANG Qian, MA Rui, CUI Yan, YU Sijiu. Molecular Mechanism of AMPK/SIRT1 Mediating Adiponectin Promoting Lactate Transport in Yak Sertoli Cells [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(7): 3495-3506. |
| [2] | NIU Yueyue, CUI Yan, YU Sijiu, HE Junfeng, YANG Shanshan, QI Zhengman, DOU Wanwan, CHEN Chunyan, DENG Yanjiang. Analysis of JAK2, STAT3, P-JAK2/STAT3 and PCNA Proteins Expression in the Lungs of Yaks of Different Ages [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(6): 2957-2967. |
| [3] | QIU Qian, SANG Rui, WANG Wei, LIU Xinman, YU Minghong, LIU Xiaotong, YU Tian, ZHANG Xuemei. Study on the Activity of Huning Powder against Chicken Lung-derived E. coli and the in vitro Effects of Anti-inflammation and Anti-oxidation [J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(4): 1969-1980. |
| [4] | Zenghua LU, Yan CUI, Sijiu YU, Xuefeng BAI, Hongqin LU, Junfeng HE, Kai LU, Guoliang ZHAI, Zhengman QI. Effect of Erythropoietin on the Expression of Apoptotic Factor in Yak Renal Interstitial Fibroblasts [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(8): 3460-3471. |
| [5] | Zhangrong PENG, Haoran SUN, Qiaoru ZHANG, Ying YANG, Hongying GUO, Tong CHANG, Hui ZHAO, Tietao ZHANG. Study on the Pattern of Intramuscular Fat Deposition and Its Influence in Flavor Quality of Sika Deer at Different Ages [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(8): 3541-3551. |
| [6] | Milan MA, Qi WANG, Qiu YAN, Tianan LI, Xingxu ZHAO, Yong ZHANG. Expression of HIG1 Hypoxia Inducible Domain Family Member 1C in Cryptorchidism of Yak and Its Regulatory Mechanism [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(7): 2983-2994. |
| [7] | Bohua LIU, Hanyu FU, Yuheng WANG, Suolangsizhu, Jiaqiang NIU, Yuhua BAO, Jiakui LI, Yefen XU. Isolation, Identification and Genome Analysis of Type B Pasteurella multocida Isolated from Yak in Tibetan Nakchu City [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(7): 3105-3118. |
| [8] | LUO Ting, HAN Zhu, XU Yefen, CAI Lin, SUOLANG Sizhu, XU Jinhua, NIU Jiaqiang. Whole Genome Sequencing and Sequence Analysis on T10 of Mycoplasma bovis Strain from Yaks in Xizang [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(5): 2154-2167. |
| [9] | HUANG Xianpeng, XING Jiayi, BAI Yuanyuan, JIANG Yuting, MA Zhiwei, FU Wei, LAN Daoliang. Cloning of Six Pluripotent Related Transcription Factors OSKMNL in Yak and Construction of Polycistron Lentiviral Vector [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(4): 1579-1591. |
| [10] | SHANG Kaiyuan, JIANG Mingfeng, GUAN Jiuqiang, AN Tianwu, ZHAO Hongwen, BAI Qin, WU Weisheng, LI Huade, XIE Rongqing, SHA Quan, LUO Xiaolin, ZHANG Xiangfei. Effects of Maternal Nutritional Regulation in Transition Period on Growth and Development, Serum Biochemistry and Immune Function of Yak Calves [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(4): 1638-1648. |
| [11] | ZUO Zizhen, WANG Haibo, CHAI Zhixin, FU Jianhui, ZHANG Xiangfei, LUO Xiaolin, ZHONG Jincheng. Effects of Rumen-protected Methionine on Meat Quality, Volatile Flavor Compounds and Fatty Acid Composition of Yak Semitendinosus [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(3): 1102-1114. |
| [12] | LIU Yanchen, ZHOU Shiying, ZHANG Yang, GAO Yang, GUAN Weijun. Isolation, Culture and Biological Characteristics Study of Holstein Bovine Lung Stem Cells [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(2): 540-551. |
| [13] | LIU Bin, WANG Meng, PAN Yangyang, WANG Jinglei, XU Gengquan. Effect of LPA on the Expression of HAS2, PTGS2 and PTX3 in Cumulus Cells of Yak (Bos grunniens) [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(2): 552-561. |
| [14] | LUO Shimei, CHEN Yunyi, LI Qisha, ZHOU Yanmei, HU Xuerou, LI Mingyue, WEI Ziyu, LI Fang, MA Xianping, YI Huashan. Effect of Bluetongue Virus Infection on Type Ⅰ Interferon Responses in Sheep Lung Microvascular Endothelial Cells [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(12): 5631-5640. |
| [15] | Juan WANG, Xueqiang YAO, Linlin LU, Jiang LI, Kairen ZHOU, Bintao ZHAI, Xuzheng ZHOU, Jiyu ZHANG. Clinical Efficacy Test of Compound Oxyclozanide Suspension on Fasciola hepatica and Nematodes of Yak [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(11): 5259-5266. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||