





Acta Veterinaria et Zootechnica Sinica ›› 2024, Vol. 55 ›› Issue (11): 4992-5002.doi: 10.11843/j.issn.0366-6964.2024.11.018
• Animal Genetics and Breeding • Previous Articles Next Articles
Zhenni LIU1,2( ), Jianjun LI1, Hai LIAN1, Xiaowen LEI1, Donghai TAN1, Qingyuan ZENG1, Di CHENG1, Yuling TIAN1, Zhiwei KONG1, Hualiang XIE1, Yunping ZHONG1,*(
), Jianjun LI1, Hai LIAN1, Xiaowen LEI1, Donghai TAN1, Qingyuan ZENG1, Di CHENG1, Yuling TIAN1, Zhiwei KONG1, Hualiang XIE1, Yunping ZHONG1,*( )
)
Received:2024-02-27
Online:2024-11-23
Published:2024-11-30
Contact:
Yunping ZHONG
E-mail:746178508@qq.com;451718467@qq.com
CLC Number:
Zhenni LIU, Jianjun LI, Hai LIAN, Xiaowen LEI, Donghai TAN, Qingyuan ZENG, Di CHENG, Yuling TIAN, Zhiwei KONG, Hualiang XIE, Yunping ZHONG. Population Evolution Analysis of Ganzhou Muscovy Duck Based on Whole Genome Resequencing[J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(11): 4992-5002.
Table 1
Analysis of heterozygosity in 8 duck populations"
| 群体Group | 观测杂合度Observed heterozygosity | 期望杂合度Eexpected heterozygosity |
| 赣州番鸭GZ | 0.057 1 | 0.062 6 |
| 天柱番鸭TZ | 0.048 3 | 0.062 1 |
| 赣西北番鸭GXB | 0.051 0 | 0.062 5 |
| 福建番鸭FJ | 0.051 9 | 0.061 0 |
| 樱桃谷鸭CV | 0.057 2 | 0.070 3 |
| 绿头鸭MD | 0.078 6 | 0.085 9 |
| 北京鸭PK | 0.063 1 | 0.072 6 |
| 法国番鸭FF | 0.062 6 | 0.052 7 |
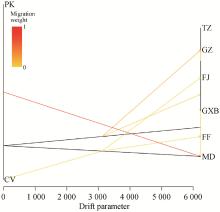
Fig. 6
Gene flow tree The black lines represent branches of the evolutionary tree; The yellow and red lines represent the migration weight, which is an indicator of the degree of gene exchange between different populations. The redder the color, the stronger the gene exchange. The scale below shows 10 times the mean standard deviation of the elements in the sample covariance matrix"

| 1 |
吴海洋, 谭东海, 谢华亮, 等. 赣南番鸭种蛋品质与孵化效果分析[J]. 江西畜牧兽医杂志, 2020, (5): 47- 48.
doi: 10.3969/j.issn.1004-2342.2020.05.016 |
|
WU H Y , TAN D H , XIE H L , et al. Analysis on egg quality and hatching effect of Gannan Muscoyduck[J]. Jiangxi Journal of Animal Husbandry & Veterinary Medicine, 2020, (5): 47- 48.
doi: 10.3969/j.issn.1004-2342.2020.05.016 |
|
| 2 |
李伟良. 清代民国时期番鸭的传入及其影响[J]. 安徽史学, 2022, (3): 58-66, 75.
doi: 10.3969/j.issn.1005-605X.2022.03.009 |
|
LI W L . Introduction of Muscovy duck and its influences during the Qing dynasty and the republic of China[J]. Historical Research in Anhui, 2022, (3): 58-66, 75.
doi: 10.3969/j.issn.1005-605X.2022.03.009 |
|
| 3 | 高超群, 曹然然, 杜文苹, 等. 基于全基因组SNP标记分析中国地方鸡品种的遗传多样性和种群结构[J]. 畜牧兽医学报, 2023, 54 (2): 554- 562. |
| GAO C Q , CAO R R , DU W P , et al. Genetic diversity and population structure analysis of Chinese native chicken breeds using genome-wide SNPs[J]. Acta Veterinaria et Zootechnica Sinica, 2023, 54 (2): 554- 562. | |
| 4 |
WEN M , WANG S Y , ZHU C C , et al. Identification of sex locus and a male-specific marker in blunt-snout bream (Megalobrama amblycephala) using a whole genome resequencing method[J]. Aquaculture, 2024, 582, 740559.
doi: 10.1016/j.aquaculture.2024.740559 |
| 5 | 章双杰, 王靖, 杨建生, 等. 长江下游地区地方鸭种全基因组序列测定及遗传多样性分析[J]. 中国家禽, 2016, 38 (23): 5- 9. |
| ZHANG S J , WANG J , YANG J S , et al. Analysis on complete genome sequencing and genetic diversity of native duck breeds in the lower reaches of Yangtze River[J]. China Poultry, 2016, 38 (23): 5- 9. | |
| 6 | 王统苗, 郭其新, 白皓, 等. 基于全基因组重测序对不同鸭遗传资源进行群体结构分析[J]. 中国畜牧杂志, 2021, 57 (11): 78- 81. |
| WANG T M , GUO Q X , BAI H , et al. Analysis of population structure of different duck genetic resources based on whole-genome resequencing[J]. Chinese Journal of Animal Science, 2021, 57 (11): 78- 81. | |
| 7 | 刘珍妮, 成笛, 孔智伟, 等. 基于全基因组重测序对大余鸭的遗传进化分析[J/OL]. 中国畜牧杂志, 2024, doi: 10.19556/j.0258-7033.20240325-02. |
| LIU Z N, CHENG D, KONG Z W, et al. Genetic evolution analysis of Dayu duck based on whole genome resequencing[J/OL]. Chinese Journal of Animal Science, 2024, doi: 10.19556/j.0258-7033.20240325-02. (in Chinese) | |
| 8 |
ZHU P Y , HE L Y , LI Y Q , et al. Correction: OTG-snpcaller: an optimized pipeline based on TMAP and GATK for SNP calling from ion torrent data[J]. PLoS One, 2015, 10 (9): e0138824.
doi: 10.1371/journal.pone.0138824 |
| 9 |
MAO J X , TIAN Y , LIU Q , et al. Revealing genetic diversity, population structure, and selection signatures of the Pacific oyster in Dalian by whole-genome resequencing[J]. Front Ecol Evol, 2024, 11, 1337980.
doi: 10.3389/fevo.2023.1337980 |
| 10 |
SENCZUK G , MACRÌ M , DI CIVITA M , et al. The demographic history and adaptation of Canarian goat breeds to environmental conditions through the use of genome-wide SNP data[J]. Genet Sel Evol, 2024, 56 (1): 2.
doi: 10.1186/s12711-023-00869-0 |
| 11 |
VANI A , KUMAR A , MAHALA S , et al. Revelation of genetic diversity and genomic footprints of adaptation in Indian pig breeds[J]. Gene, 2024, 893, 147950.
doi: 10.1016/j.gene.2023.147950 |
| 12 |
王佟, 马晓明, 余道宁, 等. 全基因组重测序解析不同牦牛类群的系谱及遗传结构[J]. 中国草食动物科学, 2023, 43 (6): 1- 6.
doi: 10.3969/j.issn.2095-3887.2023.06.001 |
|
WANG T , MA X M , YU D N , et al. Genealogy and genetic structure of different yak taxa analyzed by whole genome resequencing[J]. China Herbivore Science, 2023, 43 (6): 1- 6.
doi: 10.3969/j.issn.2095-3887.2023.06.001 |
|
| 13 |
XUE Z , WANG L , TIAN Y M , et al. A genome-wide scan to identify signatures of selection in Lueyang black-bone chicken[J]. Poult Sci, 2023, 102 (7): 102721.
doi: 10.1016/j.psj.2023.102721 |
| 14 | 马克岩, 韩金涛, 白雅琴, 等. 基于简化基因组测序的永登七山羊遗传多样性分析[J]. 畜牧兽医学报, 2023, 54 (5): 1939- 1950. |
| MA K Y , HAN J T , BAI Y Q , et al. Genetic diversity analysis of Yongdeng Qishan sheep based on specific-locus amplified fragment sequencing[J]. Acta Veterinaria et Zootechnica Sinica, 2023, 54 (5): 1939- 1950. | |
| 15 |
ASMARE S , ALEMAYEHU K , MWACHARO J , et al. Genetic diversity and within-breed variation in three indigenous Ethiopian sheep based on whole-genome analysis[J]. Heliyon, 2023, 9 (4): e14863.
doi: 10.1016/j.heliyon.2023.e14863 |
| 16 | 阿旺措吉, 仁青措姆, 黄舒泓, 等. 基于重测序数据的昌都黑山羊遗传多样性及群体结构分析[J]. 家畜生态学报, 2022, 43 (10): 30- 35. |
| AWANGCUOJI , RENQINGCUOMU , HUANG S H , et al. Genetic diversity and population structure analysis of Changdu black sheep based on resequencing data[J]. Acta Ecologae Animalis Domastici, 2022, 43 (10): 30- 35. | |
| 17 |
RAHMATALLA S A , ARENDS D , REISSMANN M , et al. Whole genome population genetics analysis of Sudanese goats identifies regions harboring genes associated with major traits[J]. BMC Genet, 2017, 18 (1): 92.
doi: 10.1186/s12863-017-0553-z |
| 18 | 鲍麒, 包鹏甲, 马晓明, 等. 肃南牦牛群体遗传结构、选择信号分析和ROH检测[J]. 中国畜牧兽医, 2022, 49 (8): 2992- 3005. |
| BAO Q , BAO P J , MA X M , et al. Genetic structure, selection signal analysis and ROH detection of Sunan yak population[J]. China Animal Husbandry & Veterinary Medicine, 2022, 49 (8): 2992- 3005. | |
| 19 |
LIU Z H , TAN X W , WANG J Y , et al. Whole genome sequencing of Luxi black head sheep for screening selection signatures associated with important traits[J]. Anim Biosci, 2022, 35 (9): 1340- 1350.
doi: 10.5713/ab.21.0533 |
| 20 | 肖倩, 张哲, 孙浩, 等. 浦东白猪遗传多样性及繁殖性能的变化分析[J]. 中国畜牧兽医, 2017, 44 (4): 1095- 1101. |
| XIAO Q , ZHANG Z , SUN H , et al. Genetic diversity and degradation analysis of reproductive performance of Pudong white pig[J]. China Animal Husbandry & Veterinary Medicine, 2017, 44 (4): 1095- 1101. | |
| 21 |
DEMENTIEVA N V , SHCHERBAKOV Y S , MITROFANOVA O V , et al. Analysis of the accumulation of homozygosity regions in chickens of the Pushkin breed using data from whole genome genotyping[J]. Ecol Genet, 2022, 20 (1): 31- 39.
doi: 10.17816/ecogen105942 |
| 22 |
童雄, 罗威, 闵力, 等. 基于全基因组SNPs分析陆丰黄牛和雷琼牛的群体结构与遗传多样性特征[J]. 中国农业科学, 2023, 56 (14): 2798- 2811.
doi: 10.3864/j.issn.0578-1752.2023.14.014 |
|
TONG X , LUO W , MIN L , et al. Population structure and genetic diversity of Lufeng cattle and Leiqiong cattle based on genome-wide SNPs[J]. Scientia Agricultura Sinica, 2023, 56 (14): 2798- 2811.
doi: 10.3864/j.issn.0578-1752.2023.14.014 |
|
| 23 | 马丽霞, 曹国伟, 朱红芳, 等. 基于RAD-seq静原鸡保种群体的遗传变异分析[J]. 畜牧兽医学报, 2022, 53 (7): 2104- 2117. |
| MA L X , CAO G W , ZHU H F , et al. Analysis of genetic variation in a conserved population of Jingyuan chickens based on RAD-seq[J]. Acta Veterinaria et Zootechnica Sinica, 2022, 53 (7): 2104- 2117. | |
| 24 |
袁娇, 徐国强, 周翔, 等. 基于SNP芯片监测通城猪的保种效果[J]. 畜牧兽医学报, 2022, 53 (8): 2514- 2523.
doi: 10.11843/j.issn.0366-6964.2022.08.010 |
|
YUAN J , XU G Q , ZHOU X , et al. SNP Chip-based monitoring of population conservation effect of Tongcheng pigs[J]. Acta Veterinaria et Zootechnica Sinica, 2022, 53 (8): 2514- 2523.
doi: 10.11843/j.issn.0366-6964.2022.08.010 |
|
| 25 |
GAO G L , ZHANG H M , NI J P , et al. Insights into genetic diversity and phenotypic variations in domestic geese through comprehensive population and pan-genome analysis[J]. J Anim Sci Biotechnol, 2023, 14 (1): 150.
doi: 10.1186/s40104-023-00944-y |
| 26 | GOLI R C , SUKHIJA N , RATHI P , et al. Unraveling the genetic tapestry of Indian chicken: a comprehensive study of molecular variations and diversity[J]. Ecol Genet Genom, 2024, 30, 100220. |
| 27 |
MEI C G , WANG H C , LIAO Q J , et al. Genetic architecture and selection of Chinese cattle revealed by whole genome resequencing[J]. Mol Biol Evol, 2018, 35 (3): 688- 699.
doi: 10.1093/molbev/msx322 |
| 28 |
WANG T , MA X M , MA C F , et al. Whole genome resequencing-based analysis of plateau adaptation in Meiren yak (Bos grunniens)[J]. Anim Biotechnol, 2024, 35 (1): 2298406.
doi: 10.1080/10495398.2023.2298406 |
| 29 | ZHI Y H , WANG D D , ZHANG K , et al. Genome-wide genetic structure of Henan indigenous chicken breeds[J]. Animals (Basel), 2023, 13 (4): 753. |
| 30 |
TANG H P , MAO S Q , XU X Y , et al. Genetic diversity analysis of different geographic populations of black carp (Mylopharyngodon piceus) based on whole genome SNP markers[J]. Aquaculture, 2024, 582, 740542.
doi: 10.1016/j.aquaculture.2024.740542 |
| 31 |
LUCEK K , GOMPERT Z , NOSIL P . The role of structural genomic variants in population differentiation and ecotype formation in Timema cristinae walking sticks[J]. Mol Ecol, 2019, 28 (6): 1224- 1237.
doi: 10.1111/mec.15016 |
| 32 |
KOMISSAROV A , VIJ S , YURCHENKO A , et al. B Chromosomes of the Asian Seabass (Lates calcarifer) contribute to genome variations at the level of individuals and populations[J]. Genes (Basel), 2018, 9 (10): 464.
doi: 10.3390/genes9100464 |
| 33 |
余道宁, 王佟, 铁中华, 等. 基于全基因组重测序对祁连白藏羊类群的群体结构分析[J]. 中国草食动物科学, 2023, 43 (6): 12- 16.
doi: 10.3969/j.issn.2095-3887.2023.06.003 |
|
YU D N , WANG T , TIE Z H , et al. Population structure analysis of Qilian white Tibetan sheep group based on whole genome resequencing[J]. China Herbivore Science, 2023, 43 (6): 12- 16.
doi: 10.3969/j.issn.2095-3887.2023.06.003 |
|
| 34 |
CHANG L L , ZHENG Y D , LI S , et al. Identification of genomic characteristics and selective signals in Guizhou black goat[J]. BMC Genomics, 2024, 25 (1): 164.
doi: 10.1186/s12864-023-09954-6 |
| 35 | 张彦威, 于丽娟, 徐新明, 等. 选择信号分析及其对绵羊功能基因的挖掘进展[J]. 中国畜牧兽医, 2023, 50 (12): 4935- 4946. |
| ZHANG Y W , YU L J , XU X M , et al. Advance on Selection signal analysis and its exploration for functional genes in sheep[J]. China Animal Husbandry & Veterinary Medicine, 2023, 50 (12): 4935- 4946. | |
| 36 | RAJAWAT D , GHILDIYAL K , SONEJITA NAYAK S , et al. Genome-wide mining of diversity and evolutionary signatures revealed selective hotspots in Indian Sahiwal cattle[J]. Gene, 2024, 901, 148178. |
| 37 | 王志武, 孙锐锋, 赵鹏, 等. SNP芯片评估欧拉藏羊群体遗传多样性和遗传结构[J]. 中国草食动物科学, 2024, 44 (1): 57- 62. |
| WANG Z W , SUN R F , ZHAO P , et al. Evaluated of genetic diversity and genetic structure in Euler Tibetan sheep population by SNPs chip[J]. China Herbivore Science, 2024, 44 (1): 57- 62. |
| [1] | Siyu LIU, Man ZHANG, Yan ZHANG, Zhitong WEI, Xinglei QI, Tengyun GAO, Xian LIU, Dong LIANG, Tong FU. Evaluation of the Conservation Effect in Nanyang Cattle Based on Resequencing Data [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(9): 3876-3886. |
| [2] | Tao ZHANG, Jiaqi LI, Lei XU, Dan WANG, Menghua ZHANG, Tao ZHANG, Mengjie YAN, Weitao WANG, Shoumin FAN, Xixia HUANG. Detection and Population Structure Analysis of Genomic Structural Variation in Xinjiang Brown Cattle Based on Whole Genome Resequencing Data [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(8): 3427-3435. |
| [3] | REN Yuwei, CHEN Xing, LIN Yanning, HUANG Xiaoxian, HONG Lingling, WANG Feng, SUN Ruiping, ZHANG Yan, LIU Hailong, ZHENG Xinli, CHAO Zhe. Investigating the Influencing Factors of Egg Laying Performance in Wenchang Chickens Based on Whole Genome Resequencing [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(2): 502-514. |
| [4] | Huili LIANG, Yujing XIE, Bowen SI, Guiying WANG, Yunliang JIANG, Guiling CAO. Analysis on Genomic Variation and Population Structure of Large-tailed Han Sheep Based on Whole Genome Resequencing [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(11): 4968-4979. |
| [5] | Xiangzhen GOU, Junxiang YANG, Zihui ZHAO, Lingxia FENG, Wanhui CHEN, Yujie LI, Zhongyu ZHANG, Keyan MA, Dongping JIANG, Rong CHANG, Yazhou WEN, Ke WANG, Youji MA. Evaluation of the Population Structure of the Ziwuling Black Goat Based on Super-GBS [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(10): 4334-4345. |
| [6] | LIN Yan, HUANG Min, LI Xiujin, ZHANG Xumeng, HUANG Yunmao, TIAN Yunbo, WU Zhongping. Uncovering Genome-wide Copy Number Variations in 8 Duck Breeds Using Whole Genome Resequencing Data [J]. Acta Veterinaria et Zootechnica Sinica, 2023, 54(9): 3700-3709. |
| [7] | LU Chang, DONG Lei, ZHANG Wanfeng, GAO Pengfei, GUO Xiaohong, CAI Chunbo, CAO Guoqing, LI Bugao. Identification and Screening of Single Nucleotide Polymorphism Loci in Jinfen White Pigs Based on Whole Genome Resequencing [J]. Acta Veterinaria et Zootechnica Sinica, 2023, 54(7): 2761-2771. |
| [8] | ZHAO Zhenjian, WANG Shujie, CHEN Dong, JI Xiang, SHEN Qi, YU Yang, CUI Shengdi, WANG Junge, CHEN Ziyang, TANG Guoqing. Population Structure and Genetic Diversity Analysis of Neijiang Pigs Based on Low-coverage Whole Genome Sequencing [J]. Acta Veterinaria et Zootechnica Sinica, 2023, 54(6): 2297-2307. |
| [9] | MA Keyan, HAN Jintao, BAI Yaqin, LI Taotao, MA Youji. Genetic Diversity Analysis of Yongdeng Qishan Sheep Based on Specific-Locus Amplified Fragment Sequencing [J]. Acta Veterinaria et Zootechnica Sinica, 2023, 54(5): 1939-1950. |
| [10] | GAO Chaoqun, CAO Ranran, DU Wenping, HU Xiaoyu, LEI Yanru, LI Wenting, KANG Xiangtao. Genetic Diversity and Population Structure Analysis of Chinese Native Chicken Breeds using Genome-wide SNPs [J]. Acta Veterinaria et Zootechnica Sinica, 2023, 54(2): 554-562. |
| [11] | HU Ziping, WANG Ligang, ZONG Wencheng, HOU Renda, SU Yanfang, NIU Naiqi, WANG Lixian, WANG Yuan, ZHNAG Longchao. Genetic Structure Analysis of Jianbai Xiang Pig Population Based on Genomic SNP and ROH [J]. Acta Veterinaria et Zootechnica Sinica, 2023, 54(10): 4117-4125. |
| [12] | YUAN Jiao, XU Guoqiang, ZHOU Xiang, XU Sanping, LI Sheng, LI Wangming, LIU Bang. SNP Chip-Based Monitoring of Population Conservation Effect of Tongcheng Pigs [J]. Acta Veterinaria et Zootechnica Sinica, 2022, 53(8): 2514-2523. |
| [13] | LIU Hongxiang, SHEN Yongjie, ZHANG Lihua, ZHANG Shuangjie, WANG Jing, ZHU Jie, CHEN Yuzhe, ZHU Chunhong, SONG Weitao, ZHANG Dan, TAO Zhiyun, XU Wenjuan, LIU Honglin, LI Huifang. Genetic Diversity Evaluation of Loumen Duck Based on Reduced-Representation Genome Sequencing [J]. Acta Veterinaria et Zootechnica Sinica, 2022, 53(6): 1735-1748. |
| [14] | LI Jing, CHENG Luguang, WAN Jiusheng, CHEN Chao, DENG Weidong, ZHANG Zhenghong, ZHANG Zhi, LI Liguang. A Study of Genetic Population Structure Analysis and Selection Signatures in Kunming Dog [J]. Acta Veterinaria et Zootechnica Sinica, 2022, 53(5): 1455-1464. |
| [15] | LIU Xiaojing, LIU Lu, WANG Jie, CUI Huanxian, ZHAO Guiping, WEN Jie. Genome-wide Association Study of Chicken Blood Glucose Traits Using Whole Genome Resequencing [J]. Acta Veterinaria et Zootechnica Sinica, 2020, 51(6): 1187-1195. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||