





Acta Veterinaria et Zootechnica Sinica ›› 2025, Vol. 56 ›› Issue (1): 246-258.doi: 10.11843/j.issn.0366-6964.2025.01.023
• Animal Biotechnology and Reproduction • Previous Articles Next Articles
NIU Yifan1,2( ), LI Chongyang1, ZHANG Peipei1, ZHANG Hang1, FENG Xiaoyi1, YU Zhou1, CAO Jianhua1, DU Weihua1, WAN Pengcheng3, MA Youji2,*(
), LI Chongyang1, ZHANG Peipei1, ZHANG Hang1, FENG Xiaoyi1, YU Zhou1, CAO Jianhua1, DU Weihua1, WAN Pengcheng3, MA Youji2,*( ), ZHAO Xueming1,*(
), ZHAO Xueming1,*( )
)
Received:2024-07-02
Online:2025-01-23
Published:2025-01-18
Contact:
MA Youji, ZHAO Xueming
E-mail:nyf.niuyifan@qq.com;yjma@gsau.edu.cn;zhaoxueming@caas.cn
CLC Number:
NIU Yifan, LI Chongyang, ZHANG Peipei, ZHANG Hang, FENG Xiaoyi, YU Zhou, CAO Jianhua, DU Weihua, WAN Pengcheng, MA Youji, ZHAO Xueming. Microamplification System Evaluation of Bovine Biopsied Embryo Cells[J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(1): 246-258.
Table 1
Summary of the quality control results of bovine biopsied embryo cell amplification products in different numbers"
| 细胞数量 Cell number | 体系名称 System name | 扩增产物浓度/(ng·μL-1) Post-amplification concentration | 体积/μL Volume | 扩增产物总质量/μg Total mass |
| 1C | MDA | 754.50±57.28A | 50 | 37.73±2.86A |
| MALBAC | 24.17±11.17B | 65 | 1.57±11.17B | |
| 5C | MDA | 941.00±174.37A | 50 | 47.05±8.72A |
| MALBAC | 29.35±0.41B | 65 | 1.91±0.02B | |
| 10C | MDA | 805.60±20.27A | 50 | 40.28±1.01A |
| MALBAC | 26.87±1.40B | 65 | 1.75±0.09B |

Fig. 2
The agarose gel electrophoresis maps of different number of bovine embryo biopsies were amplified by different systems A.MDA system: 11, 12, 13, 14 are 1C samples, 1, 2, 3, 4, 5 are 5C samples, 6, 7, 8, 9, 10 are 10C samples; B.MALBAC system: 1, 2, 3, 4, 5 are 1C samples, 6, 7, 8, 9, 10 are 5C samples, 11, 12, 13, 14, 15, 16 are 10C samples"

Table 3
Summary of resequencing data of amplified products of biopsied embryonic cells"
| 体系名称 System | 样品数量 Sample quantity | 细胞数量 Cell number | 原始测序量/G Raw date | 原始reads Raw reads | Q20/% | Q30/% | GC/% |
| NC | 1 | / | 78.08 | 240 265 726 | 97.28 | 92.27 | 42.77 |
| MDA | 4 | 1C | 190.69 | 635 613 479 | 98.30 | 94.34 | 42.37 |
| 4 | 5C | 225.79 | 752 633 841 | 98.30 | 94.23 | 42.69 | |
| 5 | 10C | 301.29 | 1 004 318 819 | 98.22 | 93.93 | 40.69 | |
| MALBAC | 3 | 1C | 159.68 | 532 248 347 | 98.27 | 94.23 | 43.43 |
| 4 | 5C | 217.24 | 724 110 484 | 98.13 | 93.82 | 43.22 | |
| 6 | 10C | 343.16 | 1 143 852 069 | 98.15 | 93.78 | 43.19 |

Fig. 3
Comparative statistical analysis map of amplified products of bovine biopsied embryos in different numbers A. The mapping rate of the amplification products of different numbers of biopsied embryo cells between the two systems; B. The correct mapping rate; C. The dup rate. The different capital letters mean extremely significant difference (P < 0.01), the different small letters mean significant difference (P < 0.05), and no letter mark mean no significant difference (P>0.05), the same as below"


Fig. 4
Statistical analysis map of sequencing depth and coverage of amplification products of bovine embryo biopsies in different numbers A. The coverage at least 5× of the amplification products of the two systems for different numbers of biopsied embryo cells; B. Coverage at least 10× of the amplification products of the two systems for different numbers of biopsied embryo cells"


Fig. 5
Statistical analysis map of sequencing and quality control of amplification products of bovine embryo biopsies in different numbers A. The typing consistent rate of amplification products of different number of biopsied embryo cells; B. Allele drop-out rate of amplification products of different number of biopsied embryo cells; C. False positive rate of amplification products of different number of biopsied embryo cells; D. 10× call rate of amplification products of different number of biopsied embryo cells"
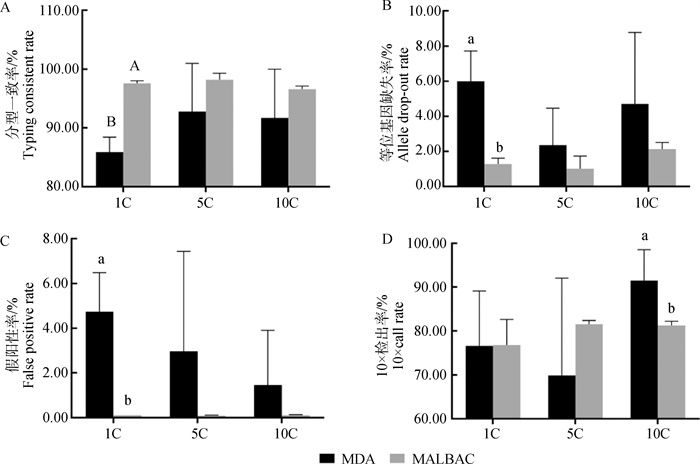

Fig. 6
Comparative statistical analysis map of amplified products of bovine biopsied embryos A. The overall mapping rate of the amplification products of the two systems; B. The correct mapping rate of the amplification products of the two systems; C. The dup rate of the amplification products of the two systems"


Fig. 8
Statistical analysis chart of data quality control indexes of amplified products of bovine biopsied embryos A. The overall typing consistent rate of the products amplified by the two systems; B. The allele drop-out rate of the products amplified by the two systems; C. The false positive rate of the products amplified by the two systems; D. The 10× call rate of the products amplified by the two systems"

Table 4
Statistical table of full-sib and half-sib loci consistency"
| 样本1 Sample1 | 样本2 Sample2 | 细胞数量 Cell number | 样本关系 Sample relationship | 分型一致率/% Consistent rate of typing | 分型一致位点数 The number of consistent loci of typing | 分型不一致位点数 The number of inconsistent loci of typing |
| Y1 | Y2 | 1C | 全同胞 | 79.93 | 15 423 363 | 3 872 311 |
| Y3 | Y4 | 5C | 77.86 | 14 664 766 | 4 169 400 | |
| Y5 | Y6 | 10C | 84.02 | 16 163 185 | 3 075 191 | |
| Q1 | Q2 | 10C | 87.82 | 23 650 146 | 3 279 974 | |
| Y2 | Y4 | 5C | 半同胞 | 76.38 | 14 269 464 | 4 413 134 |
| Y4 | Y5 | 10C | 79.28 | 14 567 703 | 3 807 469 | |
| Y2 | Y5 | 10C | 77.88 | 15 036 581 | 4 270 454 | |
| Q3 | Q4 | 5C | 82.28 | 20 365 863 | 4 385 250 | |
| Q5 | Q2 | 10C | 82.25 | 22 009 254 | 4 748 595 | |
| Q6 | Q2 | 10C | 81.84 | 21 673 120 | 4 808 018 |
| 1 |
牛一凡, 杨柏高, 张培培, 等. 牛胚胎基因组选择研究进展[J]. 畜牧兽医学报, 2023, 54 (11): 4449- 4457.
doi: 10.11843/j.issn.0366-6964.2023.11.002 |
|
NIU Y F , YANG B G , ZHANG P P , et al. Advances in bovine embryo genome selection[J]. Acta Veterinaria et Zootechnica Sinica, 2023, 54 (11): 4449- 4457.
doi: 10.11843/j.issn.0366-6964.2023.11.002 |
|
| 2 |
OLIVEIRA C S , DA SILVA M V G B , QUINTÃO C C , et al. Imputation accuracy for genomic selection using embryo biopsy samples in Gir[J]. Reprod Biol, 2023, 23 (2): 100765.
doi: 10.1016/j.repbio.2023.100765 |
| 3 | MULLAART E, WELLS D. Embryo biopsies for genomic selection[M] //NIEMANN H, WRENZYCKI C. Animal Biotechnology 2: Emerging Breeding Technologies. Cham: Springer, 2018: 81-94. |
| 4 |
OLIVEIRA C S , CAMARGO L S A , DA SILVA M V G B , et al. Embryo biopsies for genomic selection in tropical dairy cattle[J]. Anim Reprod, 2023, 20 (2): e20230064.
doi: 10.1590/1984-3143-ar2023-0064 |
| 5 |
LU N , QIAO Y , LU Z H , et al. Chimera: the spoiler in multiple displacement amplification[J]. Comput Struct Biotechnol J, 2023, 21, 1688- 1696.
doi: 10.1016/j.csbj.2023.02.034 |
| 6 |
ALMODIN C G , MORON A F , KULAY L JR , et al. A bovine protocol for training professionals in preimplantation genetic diagnosis using polymerase chain reaction[J]. Fertil Steril, 2005, 84 (4): 895- 899.
doi: 10.1016/j.fertnstert.2005.02.051 |
| 7 |
POLISSENI J , DE SÁ W F , DE OLIVEIRA GUERRA M , et al. Post-biopsy bovine embryo viability and whole genome amplification in preimplantation genetic diagnosis[J]. Fertil Steril, 2010, 93 (3): 783- 788.
doi: 10.1016/j.fertnstert.2008.10.023 |
| 8 |
GAWAD C , KOH W , QUAKE S R . Single-cell genome sequencing: current state of the science[J]. Nat Rev Genet, 2016, 17 (3): 175- 188.
doi: 10.1038/nrg.2015.16 |
| 9 |
JÄGER R . New perspectives for whole genome amplification in forensic STR analysis[J]. Int J Mol Sci, 2022, 23 (13): 7090.
doi: 10.3390/ijms23137090 |
| 10 | WANG X Y , LIU Y P , LIU H N , et al. Recent advances and application of whole genome amplification in molecular diagnosis and medicine[J]. MedComm (2020), 2022, 3 (1): e116. |
| 11 |
HUANG L , MA F , CHAPMAN A , et al. Single-cell whole-genome amplification and sequencing: methodology and applications[J]. Annu Rev Genomics Hum Genet, 2015, 16, 79- 102.
doi: 10.1146/annurev-genom-090413-025352 |
| 12 |
RAZ O , TAO L M , BIEZUNER T , et al. Whole-genome amplification-surveying yield, reproducibility, and heterozygous balance, reported by STR-targeting MIPs[J]. Int J Mol Sci, 2022, 23 (11): 6161.
doi: 10.3390/ijms23116161 |
| 13 | 姚雅馨, 喇永富, 狄冉, 等. 不同单细胞全基因组扩增方法的比较及MALBAC在辅助生殖中的应用[J]. 遗传, 2018, 40 (8): 620- 631. |
| YAO Y X , LA Y F , DI R , et al. Comparison of different single cell whole genome amplification methods and MALBAC applications in assisted reproduction[J]. Hereditas (Beijing), 2018, 40 (8): 620- 631. | |
| 14 |
ZHOU X X , XU Y , ZHU L B , et al. Comparison of multiple displacement amplification (MDA) and multiple annealing and looping-based amplification cycles (MALBAC) in limited DNA sequencing based on tube and droplet[J]. Micromachines (Basel), 2020, 11 (7): 645.
doi: 10.3390/mi11070645 |
| 15 |
DEAN F B , HOSONO S , FANG L H , et al. Comprehensive human genome amplification using multiple displacement amplification[J]. Proc Natl Acad Sci U S A, 2002, 99 (8): 5261- 5266.
doi: 10.1073/pnas.082089499 |
| 16 |
ORDÓÑEZ C D , REDREJO-RODRÍGUEZ M . DNA polymerases for whole genome amplification: considerations and future directions[J]. Int J Mol Sci, 2023, 24 (11): 9331.
doi: 10.3390/ijms24119331 |
| 17 |
HUTCHISON III C R , SMITH H O , PFANNKOCH C , et al. Cell-free cloning using φ29 DNA polymerase[J]. Proc Natl Acad Sci U S A, 2005, 102 (48): 17332- 17336.
doi: 10.1073/pnas.0508809102 |
| 18 |
PATRO S C , NIYONGABO A , MALDARELLI F , et al. New approaches to multi-parametric HIV-1 genetics using multiple displacement amplification: determining the what, how, and where of the HIV-1 reservoir[J]. Viruses, 2021, 13 (12): 2475.
doi: 10.3390/v13122475 |
| 19 |
VOLOZONOKA L , MISKOVA A , GAILITE L . Whole genome amplification in preimplantation genetic testing in the era of massively parallel sequencing[J]. Int J Mol Sci, 2022, 23 (9): 4819.
doi: 10.3390/ijms23094819 |
| 20 |
LI N , WANG L , WANG H , et al. The performance of whole genome amplification methods and next-generation sequencing for pre-implantation genetic diagnosis of chromosomal abnormalities[J]. J Genet Genomics, 2015, 42 (4): 151- 159.
doi: 10.1016/j.jgg.2015.03.001 |
| 21 |
CHEN M F , SONG P F , ZOU D , et al. Comparison of multiple displacement amplification (MDA) and multiple annealing and looping-based amplification cycles (MALBAC) in single-cell sequencing[J]. PLoS One, 2014, 9 (12): e114520.
doi: 10.1371/journal.pone.0114520 |
| 22 |
LYU L , ASGHAR U , FU J Y , et al. Comparative analysis of single-cell genome sequencing techniques toward the characterization of germline and somatic genomes in ciliated protists[J]. Eur J Protistol, 2023, 88, 125969.
doi: 10.1016/j.ejop.2023.125969 |
| 23 |
HE F , ZHOU W J , CAI R , et al. Systematic assessment of the performance of whole-genome amplification for SNP/CNV detection and β-thalassemia genotyping[J]. J Hum Genet, 2018, 63 (4): 407- 416.
doi: 10.1038/s10038-018-0411-5 |
| 24 |
LIU W Q , ZHANG H M , HU D , et al. The performance of MALBAC and MDA methods in the identification of concurrent mutations and aneuploidy screening to diagnose beta-thalassaemia disorders at the single- and multiple-cell levels[J]. J Clin Lab Anal, 2018, 32 (2): e22267.
doi: 10.1002/jcla.22267 |
| 25 |
FU Y , SHEN X T , WU H T , et al. Preimplantation genetic testing for monogenic disease of spinal muscular atrophy by multiple displacement amplification: 11 unaffected livebirths[J]. Int J Med Sci, 2019, 16 (9): 1313- 1319.
doi: 10.7150/ijms.32319 |
| 26 |
LAURI A , LAZZARI G , GALLI C , et al. Assessment of MDA efficiency for genotyping using cloned embryo biopsies[J]. Genomics, 2013, 101 (1): 24- 29.
doi: 10.1016/j.ygeno.2012.09.002 |
| 27 |
SOBOL M S , KASTER A K . Back to basics: a simplified improvement to multiple displacement amplification for microbial single-cell genomics[J]. Int J Mol Sci, 2023, 24 (5): 4270.
doi: 10.3390/ijms24054270 |
| 28 |
DE BOURCY C F A , DE VLAMINCK I , KANBAR J N , et al. A quantitative comparison of single-cell whole genome amplification methods[J]. PLoS One, 2014, 9 (8): e105585.
doi: 10.1371/journal.pone.0105585 |
| 29 |
PAN X H , URBAN A E , PALEJEV D , et al. A procedure for highly specific, sensitive, and unbiased whole-genome amplification[J]. Proc Natl Acad Sci U S A, 2008, 105 (40): 15499- 15504.
doi: 10.1073/pnas.0808028105 |
| 30 |
LASKEN R S . Single-cell sequencing in its prime[J]. Nat Biotechnol, 2013, 31 (3): 211- 212.
doi: 10.1038/nbt.2523 |
| 31 |
ZONG C H , LU S J , CHAPMAN A R , et al. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell[J]. Science, 2012, 338 (6114): 1622- 1626.
doi: 10.1126/science.1229164 |
| 32 |
CHEN C Y , XING D , TAN L Z , et al. Single-cell whole-genome analyses by Linear Amplification via Transposon Insertion (LIANTI)[J]. Science, 2017, 356 (6334): 189- 194.
doi: 10.1126/science.aak9787 |
| 33 |
ZHAO X M , HAO H S , DU W H , et al. Melatonin inhibits apoptosis and improves the developmental potential of vitrified bovine oocytes[J]. J Pineal Res, 2016, 60 (2): 132- 141.
doi: 10.1111/jpi.12290 |
| 34 |
HOU Y , WU K , SHI X L , et al. Comparison of variations detection between whole-genome amplification methods used in single-cell resequencing[J]. GigaScience, 2015, 4 (1): s13742-015-0068-3.
doi: 10.1186/s13742-015-0068-3 |
| 35 |
TREFF N R , SU J , TAO X , et al. Single-cell whole-genome amplification technique impacts the accuracy of SNP microarray-based genotyping and copy number analyses[J]. Mol Hum Reprod, 2011, 17 (6): 335- 343.
doi: 10.1093/molehr/gaq103 |
| 36 | HUMBLOT P , LE BOURHIS D , FRITZ S , et al. Reproductive technologies and genomic selection in cattle[J]. Vet Med Int, 2010, 2010, 192787. |
| 37 |
HELLANI A , COSKUN S , TBAKHI A , et al. Clinical application of multiple displacement amplification in preimplantation genetic diagnosis[J]. Reprod Biomed Online, 2005, 10 (3): 376- 380.
doi: 10.1016/S1472-6483(10)61799-3 |
| 38 |
REN Z , ZHOU C Q , XU Y W , et al. Mutation and haplotype analysis for Duchenne muscular dystrophy by single cell multiple displacement amplification[J]. Mol Hum Reprod, 2007, 13 (6): 431- 436.
doi: 10.1093/molehr/gam020 |
| 39 |
RENWICK P J , LEWIS C M , ABBS S , et al. Determination of the genetic status of cleavage-stage human embryos by microsatellite marker analysis following multiple displacement amplification[J]. Prenat Diagn, 2007, 27 (3): 206- 215.
doi: 10.1002/pd.1638 |
| 40 |
SPITS C , LE CAIGNEC C , DE RYCKE M , et al. Optimization and evaluation of single-cell whole-genome multiple displacement amplification[J]. Hum Mutat, 2006, 27 (5): 496- 503.
doi: 10.1002/humu.20324 |
| 41 |
MACARTHUR D . Face up to false positives[J]. Nature, 2012, 487 (7408): 427- 428.
doi: 10.1038/487427a |
| 42 |
YANG C H , XIAO Y , WANG X G , et al. Coordinated alternation of DNA methylation and alternative splicing of PBRM1 affect bovine sperm structure and motility[J]. Epigenetics, 2023, 18 (1): 2183339.
doi: 10.1080/15592294.2023.2183339 |
| 43 |
KENNY D , BERRY D P , PABIOU T , et al. Variation in the proportion of the segregating genome shared between full-sibling cattle and sheep[J]. Genet Sel Evol, 2023, 55 (1): 27.
doi: 10.1186/s12711-023-00802-5 |
| [1] | Yifan NIU, Chongyang LI, Baigao YANG, Peipei ZHANG, Hang ZHANG, Xiaoyi FENG, Jianhua CAO, Zhou YU, Youji MA, Xueming ZHAO. Evaluation of the Effect of Different Single Cell Whole Genome Amplification Systems on the Amplification of Bovine Trace Blood DNA [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(8): 3436-3445. |
| [2] | LI Keanning, DU Lili, AN Bingxing, DENG Tianyu, LIANG Mang, CAO Sheng, DU Yueying, XU Lingyang, GAO Xue, ZHANG Lupei, LI Junya, GAO Huijiang. Genetic Parameter Estimation and Genome-Wide Association Study for Carcass Traits and Primal Cuts Weight Traits in Huaxi Cattle [J]. Acta Veterinaria et Zootechnica Sinica, 2023, 54(9): 3664-3676. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||