





Acta Veterinaria et Zootechnica Sinica ›› 2025, Vol. 56 ›› Issue (5): 2279-2291.doi: 10.11843/j.issn.0366-6964.2025.05.026
• Preventive Veterinary Medicine • Previous Articles Next Articles
PAN Hong( ), ZHOU Saisai, YUAN Honggen, SONG Yunfeng*(
), ZHOU Saisai, YUAN Honggen, SONG Yunfeng*( )
)
Received:2024-07-04
Online:2025-05-23
Published:2025-05-27
Contact:
SONG Yunfeng
E-mail:panh@webmail.hzau.edu.cn;syf@mail.hzau.edu.cn
CLC Number:
PAN Hong, ZHOU Saisai, YUAN Honggen, SONG Yunfeng. Screening of Host Proteins for Foot and Mouth Disease Virus 3′UTR Negative-strand Interaction[J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(5): 2279-2291.
Table 1
Primer for PCR"
| 序号 No. | 引物名称 Primer name | 序列(5′→3′) Sequence |
| 1 | 3′UTR-T7-s | AATACGACTCACTATAGGGAGATCCCTCAGATGCCAC |
| 2 | 3′UTR-T7-a | AATACGACTCACTATAGGGAGATTTTTTTTTTTTTTGGAT |
| 3 | 3′UTR-HindⅢ-s | CC$\underline{{\rm{AAGCTT}}}$TCCCTCAGATGCCACTAT |
| 4 | 3′UTR-BamhⅠ-a | CGC$\underline{{\rm{GGATCC}}}$TTTTTTTTTTTTTTGGATTA |
| 5 | 5′UTR-T7-s | TAATACGACTCACTATAGGGAGAAAGTAACACCGTCGCTCCCG |
| 6 | 5′UTR-T7-a | TAATACGACTCACTATAGGGAGAAGGTTTAGTAGTGGGTAATGGAA |
| 7 | 5′UTR-s | AAGTAACACCGTCGCTCCCG |
| 8 | 5′UTR-a | AGGTTTAGTAGTGGGTAATGGAA |
| 9 | ty-3′UTR-s | CTATTGGCAACAGGCCTCTGA |
| 10 | ty-3′UTR-a | TAAGGAAGCGGGGAAAACCT |
| 11 | Ddx18-KpnⅠ-s | CGG$\underline{{\rm{GGTACC}}}$TGGCAAGATGTCGCAGTTAC |
| 12 | Ddx18-XhoⅠ-s | CCG$\underline{{\rm{CTCGAG}}}$CCTCAGTGTGAGAACTGCCTG |
| 13 | 2C-EcoRⅠ-s | CCG$\underline{{\rm{GAATTC}}}$ATGCTCAAAGCACGTGAC |
| 14 | 2C-BamHⅠ-a | CGC$\underline{{\rm{GGATCC}}}$TTGCTTGAAAATCGG |
| 15 | SL1-T7-s | TAATACGACTCACTATAGGGTCCCTCAGATGCCAC |
| 16 | SL1-a(1-51) | TACGGCGTCGCGCGC |
| 17 | SL2-T7-s | TAATACGACTCACTATAGGGGGAGTAGAAAACCGT |
| 18 | SL2-a(52-92) | GGATTAAGGAAGCGG |
Table 2
Sequence"
| 序列名称 Name | 序列(5′→3′) Sequence | 毒株信息 Strain |
| 3′UTR | TCCCTCAGATGCCACTATTGGCAACAGGCCTCTGAGGCGCGCGACGCCGTAG GAGTAGAAAACCGTAAAGGTTTTCCCCGCTTCCTTAATCCAAAAAAAAAAA AAA | O/HKN/1/2015 |
| 5′UTR | AAGTAACACCGTCGCTCCCGACGTTCAAAGGGAGGGAACCACAAGCTTGCA GCAACTTTCCCGGCGTCAACGGGATGCAACCGCAAGATGAACCTTCACCCGG AAGTAAAACGGCAACTCTACATAGTTTTGCCCGGTTTTATGAGAAACGGGA CGTCTGCGCACGAAACGCGCCGTCGCTTGAGGAAGACTTGTACAAACACGAT TTAAGCAGGTTTCCACAACTGATAAAAATCCGTGCATTTTGAAGCCTCGCC TGGTCTTTCCAGGTCTAGAGGGGCGACACTTTGTACTGTGCTCGACTCCACG CTCGGTCCACTGGCGGGTGTTAGTAGCAGCACTGTTGCTTCGTAGCGGAGC ATGGTGGCCGTGGGAACTCCTCCTTGGTGACAAGGACCCACGGGGCCGAAA GCCACGTCCGGACGGACCCACCATGTGTGCAACCCCAGCACGGCAACCTTACT GCGAACACCACCTTTAAGGTGACACTGATACTGGTACTCGGTCACTGGTGAC AGACTAAGGATGCCCTTCAGGTACCCCGAGGTAGCACGGGACACTCGGGATC TGAGAAGGGGATTGGGACTTCTTTAAAAGTGTCCAATTTAAAAAGCTTCTA TGTCTGAATAGGCGACCGGAGGCCGGCGCCTTTCCATTACCCACTACTAAACCT | O/HKN/1/2015 |
| Negative | AACTTGTTTATTGCAGCTTATAATGGTTACAAATAAAGCAATAGCATCAC AAATTTCACAAATAAAGCATTTTTTTCACTGCATTCTAGTTGTGGTTTGTC CAAACTCATCAATGTATCTTA | SV40 poly(A) signal |
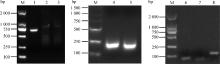
Fig. 2
In vitro transcription bands were biotin-labeled for the 5′UTR and 3′UTR M. 2000 DNA marker; lane 1. DNA-5′UTR; lane 2. RNA-bio-ss(+) 5′UTR; lane 3. RNA-bio-ss(-) 5′UTR; lane 4. RNA-bio-ss(+) 3′UTR; lane 5. RNA-bio-ss(-)3′UTR; lane 6. RNA-bio-ss(-)SL1; lane 7. RNA-bio-ss (-) SL2; lane 8. 3′UTR-△polyU"
Table 3
Mass spectrometry protein information"
| 基因名 Gene name | 蛋白质相对分子质量/u Protein mass | 特异匹配的蛋白肽段数 Unique peptide number | 覆盖率/% Coverage | 质谱鉴定得分 Protein Q-score |
| Pabpc1 | 70 625.87 | 43 | 67.61 | 128.37 |
| Msn | 67 724.89 | 37 | 68.28 | 108.18 |
| Ddx41 | 69 774.96 | 33 | 56.75 | 92.60 |
| Lmna | 74 192.70 | 31 | 40.30 | 88.19 |
| Ddx3x | 73 056.02 | 29 | 44.11 | 84.60 |
| Hspa5 | 72 377.46 | 28 | 44.27 | 84.80 |
| Abcf2 | 71 735.81 | 24 | 36.31 | 66.05 |
| Rbm14 | 69 405.98 | 22 | 28.85 | 59.61 |
| Hspa8 | 70 827.21 | 18 | 32.97 | 50.42 |
| Gtpbp4 | 74 065.87 | 16 | 31.70 | 44.23 |
| Syncrip | 69 589.61 | 15 | 25.04 | 42.10 |
| Pes1 | 67 752.98 | 15 | 23.29 | 41.00 |
| Kif2a | 79 706.57 | 13 | 18.16 | 33.84 |
| Lsg1 | 73 111.02 | 13 | 21.12 | 36.69 |
| Hspa9 | 73 415.63 | 13 | 24.45 | 35.32 |
| Ddx5 | 69 246.81 | 13 | 33.88 | 34.71 |
| Hnrnpm | 77 597.38 | 12 | 19.07 | 35.05 |
| Ezr | 69 363.63 | 12 | 33.96 | 30.06 |
| Nf2 | 69 731.98 | 11 | 21.31 | 30.28 |
| Hbs1l | 75 053.30 | 10 | 16.28 | 27.64 |
| Ddx18 | 74 134.31 | 10 | 16.36 | 27.08 |
| Ddx55 | 68 421.28 | 10 | 15.50 | 27.34 |
| Ddx17 | 72 353.99 | 10 | 28.62 | 27.66 |
Fig. 7
Verify Ddx18 protein and FMDV 3′UTR A. Dual enzyme digestion to identify pCAGGS-HA-Ddx18 eukaryotic expression vector (M. DNA marker; Lane 1.pCAGGS-HA-Ddx18); B. Western blot verification of the expression of Ddx18 protein (M.180 ku protein marker; Lane 1. Ddx18 protein); C.3′UTR negative-strand by RNA pull down of HA-Ddx18 (M.180 ku protein marker; lane 1. input; Lane 2.Blank; lane 3.Bio-ds-3′UTR RNA; Lane 4.Bio-ss(-) 3′UTR); D. Ddx18 protein by RIP pull-down of the 3′UTR negative-strand; E. Purified Ddx18 protein with His tag (M.180 ku protein marker; Lane 1.Blank; Lane 2.purified Ddx18 protein with His tag); F. Western blot verification of normal expression of the purified His-Ddx18 protein (M.180 ku protein marker; Lanes 3 and 4.Ddx18 purified protein with His tag); G. Verification of direct interaction of the 3′UTR negative-strand with His-Ddx18 protein by EMSA assay"
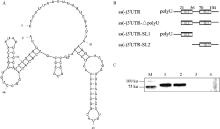
Fig. 8
3′UTR predicted structure and truncation diagram and validation of Ddx18 and 3′UTR interaction region A. Schematic diagram of single negative-strand 3′UTR RNA secondary structure; B. Schematic diagram of single negative-strand 3′UTR truncation; C. RNA pull down to probe the region of Ddx18 interactions with single negative-strand 3′UTR truncated RNA (M.180 ku protein marker; 1. Bio-ss-3′UTR; 2. Bio-ss-3′UTR-ΔpolyU; 3.Bio-SL1; 4.Bio-SL2)"
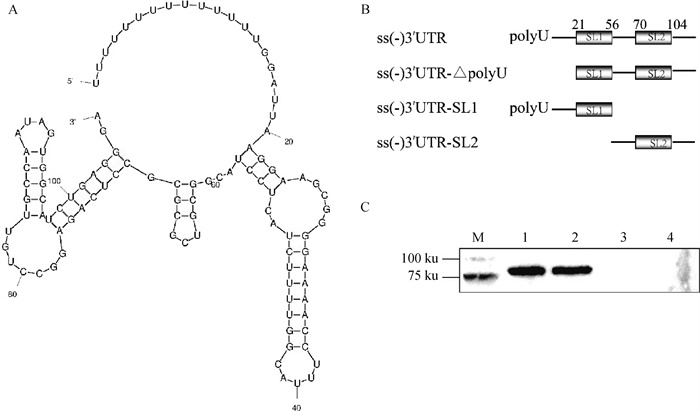
Fig. 9
Ddx18 interacts with 2C by immunoprecipitation A. PCR amplification of 2C target fragment (M. DNA relative molecular mass; Lane 1. 2C); B. Double digestion of p3×FLAG-2C plasmid (M. DNA marker; Lane 2. p3×FLAG-2C); C. Western blot verification of 2C protein expression (M.180 ku protein marker; Lane 1.2C protein with flag tag); D.Ddx18 immunoprecipitation with 2C"
| 1 | GRUBMANM,BAXTB.Foot-and-mouth disease[J].Clin Microbiol Rev,2004,4,2199-2207. |
| 2 |
ABDULLAHS,WUJ,WANGX,et al.Advances and breakthroughs in IRES-directed translation and replication of picornaviruses[J].mBio,2023,14,e0035823.
doi: 10.1128/mbio.00358-23 |
| 3 |
DORSCH-HÄSLERK,YOGOY,WIMMERE.Replication of picornaviruses. I. Evidence from in vitro RNA synthesis that poly(A) of the poliovirus genome is genetically coded[J].J Virol,1975,16,1512-1517.
doi: 10.1128/jvi.16.6.1512-1517.1975 |
| 4 | BELSHAMG.Translation and replication of FMDV RNA[J].Curr Top Microbiol Immunol,2005,288,43-70. |
| 5 |
SERRANOP,PULIDOM,SÁIZM,et al.The 3' end of the foot-and-mouth disease virus genome establishes two distinct long-range RNA-RNA interactions with the 5' end region[J].J Gen Virol,2006,87,3013-3022.
doi: 10.1099/vir.0.82059-0 |
| 6 |
GARCÍA-NUÑEZS,GISMONDIM,KÖNIGG,et al.Enhanced IRES activity by the 3'UTR element determines the virulence of FMDV isolates[J].Virology,2014,448,303-313.
doi: 10.1016/j.virol.2013.10.027 |
| 7 |
SÁIZM,GÓMEZS,MARTÍNEZ-SALASE,et al.Deletion or substitution of the aphthovirus 3'NCR abrogates infectivity and virus replication[J].J Gen Virol,2001,82,93-101.
doi: 10.1099/0022-1317-82-1-93 |
| 8 |
LOZANOG,MARTÍNEZ-SALASE.Structural insights into viral IRES-dependent translation mechanisms[J].Curr Opin Virol,2015,12,113-120.
doi: 10.1016/j.coviro.2015.04.008 |
| 9 |
BEDARDK,DAIJOGOS,SEMLERB.A nucleo-cytoplasmic SR protein functions in viral IRES-mediated translation initiation[J].EMBO J,2007,26,459-467.
doi: 10.1038/sj.emboj.7601494 |
| 10 |
PILIPENKOE,PESTOVAT,KOLUPAEVAV,et al.A cell cycle-dependent protein serves as a template-specific translation initiation factor[J].Genes Dev,2000,14,2028-2045.
doi: 10.1101/gad.14.16.2028 |
| 11 |
HARRISK,XIANGW,ALEXANDERL,et al.Interaction of poliovirus polypeptide 3CDpro with the 5' and 3' termini of the poliovirus genome. Identification of viral and cellular cofactors needed for efficient binding[J].J Biol Chem,1994,269,27004-2701.
doi: 10.1016/S0021-9258(18)47118-9 |
| 12 |
LIUY,WIMMERE,PAULA.Cis-acting RNA elements in human and animal plus-strand RNA viruses[J].Biochim Biophys Acta,2009,1789,495-517.
doi: 10.1016/j.bbagrm.2009.09.007 |
| 13 |
WAGGONERS,SARNOWP.Viral Ribonucleoprotein complex formation and nucleolar-cytoplasmic relocalization of nucleolin in poliovirus-infected cells[J].J Virol,1998,72,6699-6709.
doi: 10.1128/JVI.72.8.6699-6709.1998 |
| 14 | JIANGL,XIAOM,LIAOQ,et al.High-sensitivity profiling of SARS-CoV-2 noncoding region-host protein interactome reveals the potential regulatory role of negative-sense viral RNA[J].mSystems,2023,8,e0013523. |
| 15 |
付绍祖,李露露,张敬,等.猪源DDX56对口蹄疫病毒的复制及其对病毒诱导的RLR通路调节的研究[J].畜牧兽医学报,2019,50(9):1849-1856.
doi: 10.11843/j.issn.0366-6964.2019.09.012 |
|
FUS Z,LIL L,ZHANGJ,et al.Study on the replication of foot-and-mouth disease virus by porcine DDX56 and its regulation on virus-induced RLR pathway[J].Acta Veterinaria et Zootechnica Sinica,2019,50(9):1849-1856.
doi: 10.11843/j.issn.0366-6964.2019.09.012 |
|
| 16 | ABDULLAH S W, 解旋蛋白酶DDX23和DDX17对FMDV复制的调控[D]. 兰州: 中国农业科学院, 2021. |
| ABDULLAH S W. Regulation of FMDV replication by helicases DDX23 and DDX17[D]. Lanzhou: Chinese Academy of Agricultural Sciences, 2021. (in chinese). | |
| 17 |
ABDULLAHS,WUJ,ZHANGY,et al.DDX21, a host restriction factor of FMDV IRES-dependent translation and replication[J].Viruses,2021,13(9):1765.
doi: 10.3390/v13091765 |
| 18 |
ROEHLH,PARSLEYT,HOT,et al.Processing of a cellular polypeptide by 3CD proteinase is required for poliovirus ribonucleoprotein complex formation[J].J Virol,1997,71,578-585.
doi: 10.1128/jvi.71.1.578-585.1997 |
| 19 |
BANERJEER,ECHEVERRIA,DASGUPTAA.Poliovirus-encoded 2C polypeptide specifically binds to the 3'-terminal sequences of viral negative-strand RNA[J].J Virol,1997,71,9570-9578.
doi: 10.1128/jvi.71.12.9570-9578.1997 |
| 20 |
ROEHLH,SEMLERB.Poliovirus infection enhances the formation of two ribonucleoprotein complexes at the 3' end of viral negative-strand RNA[J].J Virol,1995,69,2954-2961.
doi: 10.1128/jvi.69.5.2954-2961.1995 |
| 21 |
BRUNNERJ,NGUYENJ,ROEHLH,et al.Functional interaction of heterogeneous nuclear ribonucleoprotein C with poliovirus RNA synthesis initiation complexes[J].J Virol,2005,79,3254-3266.
doi: 10.1128/JVI.79.6.3254-3266.2005 |
| 22 |
DMITRIEVAT,SHCHEGLOVAM,AGOLV.Inhibition of activity of encephalomyocarditis virus-induced RNA polymerase by antibodies against cellular components[J].Virology,1979,92,271-277.
doi: 10.1016/0042-6822(79)90131-4 |
| 23 | BROWND,CORNELLC,TRANG,et al.An authentic 3'noncoding region is necessary for efficient poliovirus replication[J].J Virol,2005,79,11962-11973. |
| 24 | BELOVG,EVSTAFIEVAA,RUBTSOVY,et al.Early alteration of nucleocytoplasmic traffic induced by some RNA viruses[J].Virology,2000,275,244-248. |
| [1] | DAI Yuxing,SHI Yinyin,WEN Zuochen,LUO Yunyan,ZHU Xueli,ZHENG Chunting,LI Shuying,HONG Liang,ZHANG Jianbin,GUO Liang,PU Lei. The Effect of Hlcs Interference on Glycolytic Gene Expression in C2C12 Cells after Myogenic Lipogenic Differentiation [J]. Acta Veterinaria et Zootechnica Sinica, 2022, 53(10): 3391-3402. |
| [2] | YANG Tao, XU Houqiang, CHEN Wei, ZHOU Di, WANG Yuanyuan, ZHU Xiaofeng. Study on Interaction between MSTN Promoter and MEF2C Transcription Factor in Cattle [J]. ACTA VETERINARIA ET ZOOTECHNICA SINICA, 2019, 50(8): 1567-1575. |
| [3] | CHEN Xiangxing, LI Jiaolong, XING Tong, ZHANG Lin, GAO Feng. Effects of H2O2 on Oxidative Damage of C2C12 Cells [J]. ACTA VETERINARIA ET ZOOTECHNICA SINICA, 2019, 50(5): 1016-1025. |
| [4] | LIU Yan,LIANG Hui-huang,LIU Yu-lan,TANG Zhong-lin,WANG Wen-jun,ZHANG Jing. LPS Induces MuRF1 Transcription through AKT/FOXO1 Mediating Pathway in C2C12 Myotubes [J]. ACTA VETERINARIA ET ZOOTECHNICA SINICA, 2016, 47(2): 374-380. |
| [5] | HUAN Cong-cong,XU Hou-qiang,CHEN Wei,CHEN Xiang,ZHAO Jia-fu,ZHANG Wen,ZHOU Di,XIA Dan. Construction of MyoD I Promoter Eukaryotic Expression Vector in Guanling Cattle and Its Expression in Mouse C2C12 Cells [J]. ACTA VETERINARIA ET ZOOTECHNICA SINICA, 2015, 46(5): 719-727. |
| [6] | WANG Guo-qing,ZHU Zi-xiang,CAO Wei-jun,YANG Fan,MAO Ruo-qing,LI Dan,LIU Lei,ZHENG Hai-xue . Construction of Porcine RIG-I Eukaryotic Expressing Plasmid and Its Antiviral Effects Research against Foot and mouth Disease Virus [J]. ACTA VETERINARIA ET ZOOTECHNICA SINICA, 2015, 46(4): 600-607. |
| [7] | ZHANG Xiaoli;;LU Zengjun;MA Xiaojun;CAO Yimei;FU Yuanfang;LIU Zaixin;XIE Qingge . Expression of NonStructural Protein 2C of Footandmouth Disease Virus in Insect Cells and Its Application [J]. ACTA VETERINARIA ET ZOOTECHNICA SINICA, 2011, 42(12): 1724-1731. |
| [8] | FU Yuan-fang;LU Zeng-jun;TIAN Mei-na;ZHANG Xiao-li;LIU Zai-xin;CAI Xue-peng. Expression of Major B-cell Epitopes within 2C Non-structural Protein of FMDV and Its Bioactivity [J]. ACTA VETERINARIA ET ZOOTECHNICA SINICA, 2008, 39(9): 1235-1239. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||