





畜牧兽医学报 ›› 2025, Vol. 56 ›› Issue (5): 2279-2291.doi: 10.11843/j.issn.0366-6964.2025.05.026
收稿日期:2024-07-04
出版日期:2025-05-23
发布日期:2025-05-27
通讯作者:
宋云峰
E-mail:panh@webmail.hzau.edu.cn;syf@mail.hzau.edu.cn
作者简介:潘红(1999-),女,壮族,广西上林人,硕士生,主要从事口蹄疫病毒非编码基因的研究,E-mail: panh@webmail.hzau.edu.cn
基金资助:
PAN Hong( ), ZHOU Saisai, YUAN Honggen, SONG Yunfeng*(
), ZHOU Saisai, YUAN Honggen, SONG Yunfeng*( )
)
Received:2024-07-04
Online:2025-05-23
Published:2025-05-27
Contact:
SONG Yunfeng
E-mail:panh@webmail.hzau.edu.cn;syf@mail.hzau.edu.cn
摘要:
本研究旨在筛选口蹄疫病毒(foot and mouth disease,FMDV)负链RNA复制过程中与3′UTR结合的宿主蛋白。首先,通过RNA pull down结合质谱,鉴定出与3′UTR负链结合的宿主蛋白。对质谱鉴定到的蛋白结果进行GO功能注释分析、KEGG通路富集分析和互作网络筛选分析,并构建真核表达质粒以及原核表达质粒,表达该互作蛋白来验证RNA与蛋白的互作特异性。其次,截短3′UTR探究RNA与蛋白的互作区域,最后利用Co-IP探究宿主互作蛋白与病毒蛋白的互作关系。结果发现与3′UTR互作结合的蛋白多为核仁蛋白,其中以DEAD-box RNA解旋酶蛋白家族的Ddx18蛋白作为重点研究对象。通过构建该蛋白的真核表达载体,利用RNA pull down、RIP, 以及EMSA证实Ddx18确实与3′UTR负链互作。转录出截短的3′UTR负链RNA,利用RNA pull down试验得出Ddx18与3′UTR负链的互作不依赖poly(U),且不与截短后的SL1和SL2互作。利用Co-IP试验证明2C病毒蛋白能与Ddx18蛋白互作。综上,3′UTR与Ddx18蛋白的互作可能基于RNA的空间结构,而2C病毒蛋白与Ddx18蛋白结合,提示2C可能间接结合3′UTR而共同形成复制复合物参与到FMDV的复制中。通过筛选与3′UTR负链互作的宿主蛋白,为进一步探究FMDV复制相关机制从而研究相关药物靶点奠定实验基础。
中图分类号:
潘红, 周赛赛, 袁红根, 宋云峰. 口蹄疫病毒3′UTR负链互作的宿主蛋白筛选[J]. 畜牧兽医学报, 2025, 56(5): 2279-2291.
PAN Hong, ZHOU Saisai, YUAN Honggen, SONG Yunfeng. Screening of Host Proteins for Foot and Mouth Disease Virus 3′UTR Negative-strand Interaction[J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(5): 2279-2291.
表 1
PCR引物序列"
| 序号 No. | 引物名称 Primer name | 序列(5′→3′) Sequence |
| 1 | 3′UTR-T7-s | AATACGACTCACTATAGGGAGATCCCTCAGATGCCAC |
| 2 | 3′UTR-T7-a | AATACGACTCACTATAGGGAGATTTTTTTTTTTTTTGGAT |
| 3 | 3′UTR-HindⅢ-s | CC$\underline{{\rm{AAGCTT}}}$TCCCTCAGATGCCACTAT |
| 4 | 3′UTR-BamhⅠ-a | CGC$\underline{{\rm{GGATCC}}}$TTTTTTTTTTTTTTGGATTA |
| 5 | 5′UTR-T7-s | TAATACGACTCACTATAGGGAGAAAGTAACACCGTCGCTCCCG |
| 6 | 5′UTR-T7-a | TAATACGACTCACTATAGGGAGAAGGTTTAGTAGTGGGTAATGGAA |
| 7 | 5′UTR-s | AAGTAACACCGTCGCTCCCG |
| 8 | 5′UTR-a | AGGTTTAGTAGTGGGTAATGGAA |
| 9 | ty-3′UTR-s | CTATTGGCAACAGGCCTCTGA |
| 10 | ty-3′UTR-a | TAAGGAAGCGGGGAAAACCT |
| 11 | Ddx18-KpnⅠ-s | CGG$\underline{{\rm{GGTACC}}}$TGGCAAGATGTCGCAGTTAC |
| 12 | Ddx18-XhoⅠ-s | CCG$\underline{{\rm{CTCGAG}}}$CCTCAGTGTGAGAACTGCCTG |
| 13 | 2C-EcoRⅠ-s | CCG$\underline{{\rm{GAATTC}}}$ATGCTCAAAGCACGTGAC |
| 14 | 2C-BamHⅠ-a | CGC$\underline{{\rm{GGATCC}}}$TTGCTTGAAAATCGG |
| 15 | SL1-T7-s | TAATACGACTCACTATAGGGTCCCTCAGATGCCAC |
| 16 | SL1-a(1-51) | TACGGCGTCGCGCGC |
| 17 | SL2-T7-s | TAATACGACTCACTATAGGGGGAGTAGAAAACCGT |
| 18 | SL2-a(52-92) | GGATTAAGGAAGCGG |
表 2
序列信息"
| 序列名称 Name | 序列(5′→3′) Sequence | 毒株信息 Strain |
| 3′UTR | TCCCTCAGATGCCACTATTGGCAACAGGCCTCTGAGGCGCGCGACGCCGTAG GAGTAGAAAACCGTAAAGGTTTTCCCCGCTTCCTTAATCCAAAAAAAAAAA AAA | O/HKN/1/2015 |
| 5′UTR | AAGTAACACCGTCGCTCCCGACGTTCAAAGGGAGGGAACCACAAGCTTGCA GCAACTTTCCCGGCGTCAACGGGATGCAACCGCAAGATGAACCTTCACCCGG AAGTAAAACGGCAACTCTACATAGTTTTGCCCGGTTTTATGAGAAACGGGA CGTCTGCGCACGAAACGCGCCGTCGCTTGAGGAAGACTTGTACAAACACGAT TTAAGCAGGTTTCCACAACTGATAAAAATCCGTGCATTTTGAAGCCTCGCC TGGTCTTTCCAGGTCTAGAGGGGCGACACTTTGTACTGTGCTCGACTCCACG CTCGGTCCACTGGCGGGTGTTAGTAGCAGCACTGTTGCTTCGTAGCGGAGC ATGGTGGCCGTGGGAACTCCTCCTTGGTGACAAGGACCCACGGGGCCGAAA GCCACGTCCGGACGGACCCACCATGTGTGCAACCCCAGCACGGCAACCTTACT GCGAACACCACCTTTAAGGTGACACTGATACTGGTACTCGGTCACTGGTGAC AGACTAAGGATGCCCTTCAGGTACCCCGAGGTAGCACGGGACACTCGGGATC TGAGAAGGGGATTGGGACTTCTTTAAAAGTGTCCAATTTAAAAAGCTTCTA TGTCTGAATAGGCGACCGGAGGCCGGCGCCTTTCCATTACCCACTACTAAACCT | O/HKN/1/2015 |
| Negative | AACTTGTTTATTGCAGCTTATAATGGTTACAAATAAAGCAATAGCATCAC AAATTTCACAAATAAAGCATTTTTTTCACTGCATTCTAGTTGTGGTTTGTC CAAACTCATCAATGTATCTTA | SV40 poly(A) signal |
表 3
质谱蛋白信息"
| 基因名 Gene name | 蛋白质相对分子质量/u Protein mass | 特异匹配的蛋白肽段数 Unique peptide number | 覆盖率/% Coverage | 质谱鉴定得分 Protein Q-score |
| Pabpc1 | 70 625.87 | 43 | 67.61 | 128.37 |
| Msn | 67 724.89 | 37 | 68.28 | 108.18 |
| Ddx41 | 69 774.96 | 33 | 56.75 | 92.60 |
| Lmna | 74 192.70 | 31 | 40.30 | 88.19 |
| Ddx3x | 73 056.02 | 29 | 44.11 | 84.60 |
| Hspa5 | 72 377.46 | 28 | 44.27 | 84.80 |
| Abcf2 | 71 735.81 | 24 | 36.31 | 66.05 |
| Rbm14 | 69 405.98 | 22 | 28.85 | 59.61 |
| Hspa8 | 70 827.21 | 18 | 32.97 | 50.42 |
| Gtpbp4 | 74 065.87 | 16 | 31.70 | 44.23 |
| Syncrip | 69 589.61 | 15 | 25.04 | 42.10 |
| Pes1 | 67 752.98 | 15 | 23.29 | 41.00 |
| Kif2a | 79 706.57 | 13 | 18.16 | 33.84 |
| Lsg1 | 73 111.02 | 13 | 21.12 | 36.69 |
| Hspa9 | 73 415.63 | 13 | 24.45 | 35.32 |
| Ddx5 | 69 246.81 | 13 | 33.88 | 34.71 |
| Hnrnpm | 77 597.38 | 12 | 19.07 | 35.05 |
| Ezr | 69 363.63 | 12 | 33.96 | 30.06 |
| Nf2 | 69 731.98 | 11 | 21.31 | 30.28 |
| Hbs1l | 75 053.30 | 10 | 16.28 | 27.64 |
| Ddx18 | 74 134.31 | 10 | 16.36 | 27.08 |
| Ddx55 | 68 421.28 | 10 | 15.50 | 27.34 |
| Ddx17 | 72 353.99 | 10 | 28.62 | 27.66 |
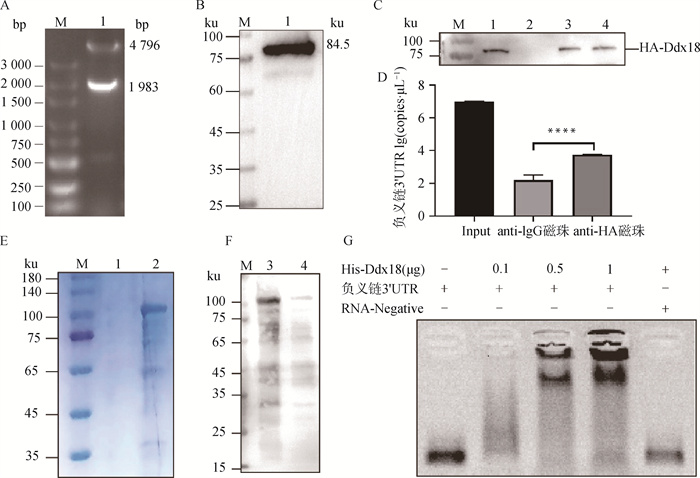
图 7
验证Ddx18蛋白与FMDV 3′UTR互作 A. 双酶切鉴定pCAGGS-HA-Ddx18真核表达载体(M. DNA相对分子质量标准;泳道1.pCAGGS-HA-Ddx18);B. Western blot验证Ddx18蛋白的表达(M. 180 ku蛋白相对分子质量标准;泳道1.Ddx18蛋白);C. 3′UTR负链通过RNA pull down结合HA-Ddx18(M. 180 ku蛋白相对分子质量标准;泳道1.Input;泳道2.Blank;泳道3.Bio-ds-3′UTR RNA;泳道4.Bio-ss(-)3′UTR);D. Ddx18蛋白通过RIP结合3′UTR负链;E. 纯化带His标签的Ddx18蛋白(M. 180 ku蛋白相对分子质量标准;泳道1.空白;泳道2.带His标签的Ddx18纯化蛋白);F. Western blot验证纯化的His-Ddx18蛋白是否正常表达(M. 180 ku蛋白相对分子质量标准;泳道3和4.带His标签的Ddx18纯化蛋白);G.通过EMSA试验验证3′UTR负链与His-Ddx18蛋白直接互作"
| 1 | GRUBMANM,BAXTB.Foot-and-mouth disease[J].Clin Microbiol Rev,2004,4,2199-2207. |
| 2 |
ABDULLAHS,WUJ,WANGX,et al.Advances and breakthroughs in IRES-directed translation and replication of picornaviruses[J].mBio,2023,14,e0035823.
doi: 10.1128/mbio.00358-23 |
| 3 |
DORSCH-HÄSLERK,YOGOY,WIMMERE.Replication of picornaviruses. I. Evidence from in vitro RNA synthesis that poly(A) of the poliovirus genome is genetically coded[J].J Virol,1975,16,1512-1517.
doi: 10.1128/jvi.16.6.1512-1517.1975 |
| 4 | BELSHAMG.Translation and replication of FMDV RNA[J].Curr Top Microbiol Immunol,2005,288,43-70. |
| 5 |
SERRANOP,PULIDOM,SÁIZM,et al.The 3' end of the foot-and-mouth disease virus genome establishes two distinct long-range RNA-RNA interactions with the 5' end region[J].J Gen Virol,2006,87,3013-3022.
doi: 10.1099/vir.0.82059-0 |
| 6 |
GARCÍA-NUÑEZS,GISMONDIM,KÖNIGG,et al.Enhanced IRES activity by the 3'UTR element determines the virulence of FMDV isolates[J].Virology,2014,448,303-313.
doi: 10.1016/j.virol.2013.10.027 |
| 7 |
SÁIZM,GÓMEZS,MARTÍNEZ-SALASE,et al.Deletion or substitution of the aphthovirus 3'NCR abrogates infectivity and virus replication[J].J Gen Virol,2001,82,93-101.
doi: 10.1099/0022-1317-82-1-93 |
| 8 |
LOZANOG,MARTÍNEZ-SALASE.Structural insights into viral IRES-dependent translation mechanisms[J].Curr Opin Virol,2015,12,113-120.
doi: 10.1016/j.coviro.2015.04.008 |
| 9 |
BEDARDK,DAIJOGOS,SEMLERB.A nucleo-cytoplasmic SR protein functions in viral IRES-mediated translation initiation[J].EMBO J,2007,26,459-467.
doi: 10.1038/sj.emboj.7601494 |
| 10 |
PILIPENKOE,PESTOVAT,KOLUPAEVAV,et al.A cell cycle-dependent protein serves as a template-specific translation initiation factor[J].Genes Dev,2000,14,2028-2045.
doi: 10.1101/gad.14.16.2028 |
| 11 |
HARRISK,XIANGW,ALEXANDERL,et al.Interaction of poliovirus polypeptide 3CDpro with the 5' and 3' termini of the poliovirus genome. Identification of viral and cellular cofactors needed for efficient binding[J].J Biol Chem,1994,269,27004-2701.
doi: 10.1016/S0021-9258(18)47118-9 |
| 12 |
LIUY,WIMMERE,PAULA.Cis-acting RNA elements in human and animal plus-strand RNA viruses[J].Biochim Biophys Acta,2009,1789,495-517.
doi: 10.1016/j.bbagrm.2009.09.007 |
| 13 |
WAGGONERS,SARNOWP.Viral Ribonucleoprotein complex formation and nucleolar-cytoplasmic relocalization of nucleolin in poliovirus-infected cells[J].J Virol,1998,72,6699-6709.
doi: 10.1128/JVI.72.8.6699-6709.1998 |
| 14 | JIANGL,XIAOM,LIAOQ,et al.High-sensitivity profiling of SARS-CoV-2 noncoding region-host protein interactome reveals the potential regulatory role of negative-sense viral RNA[J].mSystems,2023,8,e0013523. |
| 15 |
付绍祖,李露露,张敬,等.猪源DDX56对口蹄疫病毒的复制及其对病毒诱导的RLR通路调节的研究[J].畜牧兽医学报,2019,50(9):1849-1856.
doi: 10.11843/j.issn.0366-6964.2019.09.012 |
|
FUS Z,LIL L,ZHANGJ,et al.Study on the replication of foot-and-mouth disease virus by porcine DDX56 and its regulation on virus-induced RLR pathway[J].Acta Veterinaria et Zootechnica Sinica,2019,50(9):1849-1856.
doi: 10.11843/j.issn.0366-6964.2019.09.012 |
|
| 16 | ABDULLAH S W, 解旋蛋白酶DDX23和DDX17对FMDV复制的调控[D]. 兰州: 中国农业科学院, 2021. |
| ABDULLAH S W. Regulation of FMDV replication by helicases DDX23 and DDX17[D]. Lanzhou: Chinese Academy of Agricultural Sciences, 2021. (in chinese). | |
| 17 |
ABDULLAHS,WUJ,ZHANGY,et al.DDX21, a host restriction factor of FMDV IRES-dependent translation and replication[J].Viruses,2021,13(9):1765.
doi: 10.3390/v13091765 |
| 18 |
ROEHLH,PARSLEYT,HOT,et al.Processing of a cellular polypeptide by 3CD proteinase is required for poliovirus ribonucleoprotein complex formation[J].J Virol,1997,71,578-585.
doi: 10.1128/jvi.71.1.578-585.1997 |
| 19 |
BANERJEER,ECHEVERRIA,DASGUPTAA.Poliovirus-encoded 2C polypeptide specifically binds to the 3'-terminal sequences of viral negative-strand RNA[J].J Virol,1997,71,9570-9578.
doi: 10.1128/jvi.71.12.9570-9578.1997 |
| 20 |
ROEHLH,SEMLERB.Poliovirus infection enhances the formation of two ribonucleoprotein complexes at the 3' end of viral negative-strand RNA[J].J Virol,1995,69,2954-2961.
doi: 10.1128/jvi.69.5.2954-2961.1995 |
| 21 |
BRUNNERJ,NGUYENJ,ROEHLH,et al.Functional interaction of heterogeneous nuclear ribonucleoprotein C with poliovirus RNA synthesis initiation complexes[J].J Virol,2005,79,3254-3266.
doi: 10.1128/JVI.79.6.3254-3266.2005 |
| 22 |
DMITRIEVAT,SHCHEGLOVAM,AGOLV.Inhibition of activity of encephalomyocarditis virus-induced RNA polymerase by antibodies against cellular components[J].Virology,1979,92,271-277.
doi: 10.1016/0042-6822(79)90131-4 |
| 23 | BROWND,CORNELLC,TRANG,et al.An authentic 3'noncoding region is necessary for efficient poliovirus replication[J].J Virol,2005,79,11962-11973. |
| 24 | BELOVG,EVSTAFIEVAA,RUBTSOVY,et al.Early alteration of nucleocytoplasmic traffic induced by some RNA viruses[J].Virology,2000,275,244-248. |
| [1] | 廖焕程, 石正旺, 罗俊聪, 王婉莹, 冯露, 周静, 张帆, 石鑫泰, 田宏. O型口蹄疫Cathay拓扑型病毒单抗制备及双抗体夹心ELISA方法的初步建立[J]. 畜牧兽医学报, 2024, 55(9): 4012-4020. |
| [2] | 王家丽, 杨帆, 邵文华, 黄梦瑶, 曹伟军, 蒲秀瑛, 张伟, 郑海学. Tollip敲除猪肾细胞系的构建[J]. 畜牧兽医学报, 2024, 55(4): 1810-1818. |
| [3] | 董开恒, 黄书伦, 李凤娟, 李坤, 刘果, 张强, 包慧芳, 李洪炫, 卢曾军, 张小丽. 口蹄疫病毒Asia1型猪源中和抗体的筛选与抗原表位鉴定[J]. 畜牧兽医学报, 2024, 55(11): 5211-5221. |
| [4] | 李平花, 黄书伦, 张克强, 刘锋, 孙普, 李冬, 包慧芳, 曹轶梅, 白兴文, 马雪青, 李坤, 袁红, 刘在新, 卢曾军. 嵌合流行毒株G-H环基因重组口蹄疫病毒的拯救及其免疫原性分析[J]. 畜牧兽医学报, 2024, 55(11): 5222-5229. |
| [5] | 周广青, 刘晓庆, 史喜绢, 杨大鹏, 袁莉刚, 常惠芸. 口蹄疫病毒抗体SA-ELISA快速检测方法的建立[J]. 畜牧兽医学报, 2023, 54(5): 2020-2029. |
| [6] | 李硕, 张韵, 白满元, 赵瑞翀, 宋河涛, 穆素雨, 滕志东, 董虎, 马娥宁, 孙世琪, 郭慧琛, 尹双辉. 生物矿化的口蹄疫病毒样颗粒疫苗的免疫原性评价[J]. 畜牧兽医学报, 2023, 54(4): 1598-1607. |
| [7] | 郭紫晶, 陈飞, 张志雄, 柏玲, 张志东, 李彦敏. 白细胞介素-10对口蹄疫病毒感染小鼠T细胞增殖及其表达TNF-α、IFN-γ和IL-2的影响[J]. 畜牧兽医学报, 2023, 54(2): 694-705. |
| [8] | 陈文哲, 张向乐, 顾峰幸, 赵振翔, 李康丽, 薛钊宁, 郑海学, 张小丽, 朱紫祥. 猪GRK2蛋白抗口蹄疫病毒作用分析[J]. 畜牧兽医学报, 2023, 54(10): 4350-4361. |
| [9] | 韩伟建, 张俊娟, 张义明, 王家鑫, 李丽敏. 骨髓源肥大细胞通过甘露糖受体识别FMDV-VLPs的细胞因子应答[J]. 畜牧兽医学报, 2022, 53(11): 3917-3926. |
| [10] | 代宇星, 史银银, 温作晨, 罗云燕, 祝雪丽, 郑春婷, 李淑英, 洪亮, 张建斌, 郭亮, 蒲蕾. Hlcs干扰对C2C12细胞成肌成脂分化后糖酵解基因表达的影响[J]. 畜牧兽医学报, 2022, 53(10): 3391-3402. |
| [11] | 张志东, Eoin Ryan, 李彦敏. 口蹄疫病毒受体整联蛋白在绵羊不同发育阶段组织中mRNA转录水平的分析[J]. 畜牧兽医学报, 2021, 52(9): 2626-2632. |
| [12] | 李昕, 孙燕燕, 林密, 陈夏辉, 李峰松, 包艳芳, 杨光, 蒋韬. O、A、Asia 1型口蹄疫病毒胶体金定量检测免疫层析方法的建立[J]. 畜牧兽医学报, 2021, 52(4): 1042-1052. |
| [13] | 李显, 张中旺, 张富东, 吕建亮, 李嘉豪, 潘丽. 甘露糖修饰的壳聚糖聚乳酸-羟基乙酸共聚物纳米微球作为口蹄疫病毒核酸疫苗递送载体的特性评价[J]. 畜牧兽医学报, 2021, 52(12): 3557-3568. |
| [14] | 李露露, 张敬, 李丹, 郑海学. 口蹄疫病毒3D蛋白促进TLR3介导的Ⅰ型干扰素产生[J]. 畜牧兽医学报, 2020, 51(8): 1923-1931. |
| [15] | 闫鸣昊, 郝军红, 张大俊, 申超超, 徐国伟, 侯景, 张克山, 郑海学, 刘湘涛. PK-15细胞的TPL2基因敲除有利于口蹄疫病毒和塞内卡病毒复制[J]. 畜牧兽医学报, 2020, 51(5): 1060-1073. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
