





Acta Veterinaria et Zootechnica Sinica ›› 2025, Vol. 56 ›› Issue (8): 3734-3748.doi: 10.11843/j.issn.0366-6964.2025.08.016
• Animal Genetics and Breeding • Previous Articles Next Articles
CHI Shunshun1,2,3( ), WU Dan4(
), WU Dan4( ), WANG Nan2,3, WANG Wanjie3, NIE Yuxin3, MU Yulian3, LIU Zhiguo3,*(
), WANG Nan2,3, WANG Wanjie3, NIE Yuxin3, MU Yulian3, LIU Zhiguo3,*( ), ZHU Zhendong1,*(
), ZHU Zhendong1,*( ), LI Kui2,3
), LI Kui2,3
Received:2025-02-13
Online:2025-08-23
Published:2025-08-28
Contact:
LIU Zhiguo, ZHU Zhendong
E-mail:chishunshun@163.com;190833173@qq.com;liuzhiguo@caas.cn;zzd2020@qau.edu.cn
CLC Number:
CHI Shunshun, WU Dan, WANG Nan, WANG Wanjie, NIE Yuxin, MU Yulian, LIU Zhiguo, ZHU Zhendong, LI Kui. Establishment and Application of A Detection Method for MSTN Gene-Edited Pigs Based on RPA-CRISPR/Cas12a[J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(8): 3734-3748.
Table 2
crRNA sequence"
| 名称Name | 序列(5′→3′) Sequence |
| GEcrRNA1 | UAAUUUCUACUAAGUGUAGAUACUUUUGGAUGGGACUGGAUUAU |
| GEcrRNA2 | UAAUUUCUACUAAGUGUAGAUACUUUUGGAUGGGACUGGAU |
| WTcrRNA1 | UAAUUUCUACUAAGUGUAGAUAAGCUUUUGGAUGGGACUGG |
| WTcrRNA2 | UAAUUUCUACUAAGUGUAGAUAAAAUCCACAGUUAGAGGGU |
| WTcrRNA3 | UAAUUUCUACUAAGUGUAGAUAAAUCCACAGUUAGAGGGUA |
| WTcrRNA4 | UAAUUUCUACUAAGUGUAGAUAGCUUUUGGAUGGGACUGGA |
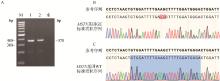
Fig. 1
Construction of standard plasmids A. PCR amplification of the MSTN gene: Marker.1 000 bp DNA marker; 1. Ear sample from MSTN gene-edited homozygous pig; 2. Ear sample from MSTN wild-type pig; H2O. Negative control without PCR template; The size of the PCR product is approximately 370 bp. B. Sanger sequencing results of the GE standard plasmid. C. Sanger sequencing results of the WT standard plasmid"
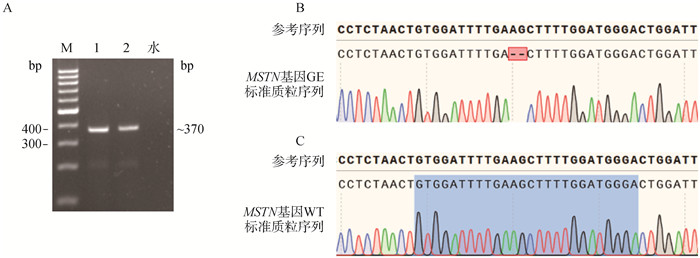
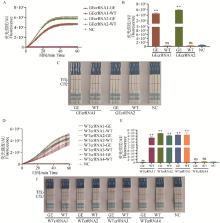
Fig. 2
crRNA screening results A-C. The fluorescence curve, fluorescence intensity bar chart, and results of colloidal gold test strips for GEcrRNA are presented in sequence; D-F. The fluorescence curve, fluorescence intensity bar chart, and results of colloidal gold test strips for WTcrRNA are presented in sequence; NC. Negative control group without crRNA. ** indicates a highly significant difference compared with the NC group (P < 0.01); ns indicates no significant difference compared with the NC group (P>0.05), the same as below. GE represents the GE standard plasmid samples, WT represents the WT standard plasmid samples; T line represents the test line, C line represents the control line; A positive result is indicated by the appearance of color on the T line alone or on both the T and C lines; A negative result is indicated by the appearance of color on the C line alone, the same as below"
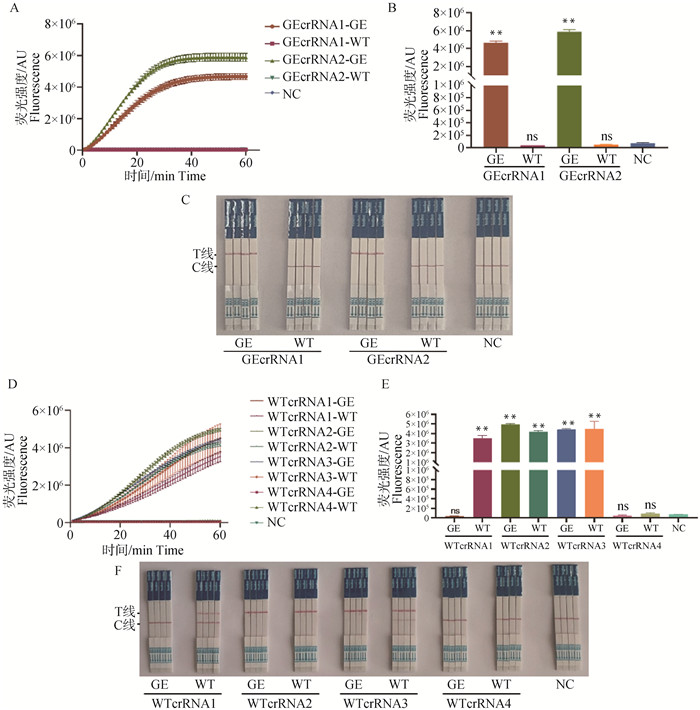
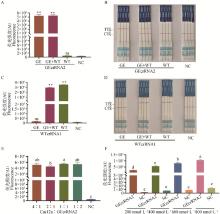
Fig. 3
Cas12a system specificity test and system optimization A, B. Fluorescence reporter system and colloidal gold test strip results for the specificity test of GEcrRNA2; C, D. Fluorescence reporter system and colloidal gold test strip results for the specificity test of WTcrRNA1; E. Fluorescence intensity of the CRISPR/Cas12a system at different molar ratios of Cas12a protein to GEcrRNA2; F. Fluorescence intensity of the CRISPR/Cas12a system at different concentrations of ssDNA probe; NC. Negative control without crRNA. GE+WT represents a 1∶1 mixture of GE standard plasmid and WT standard plasmid, the same as below. In panels E and F, different lowercase letters indicate a significant difference between groups (P < 0.05); The same lowercase letter indicates no significant difference between groups (P > 0.05), the same as below"
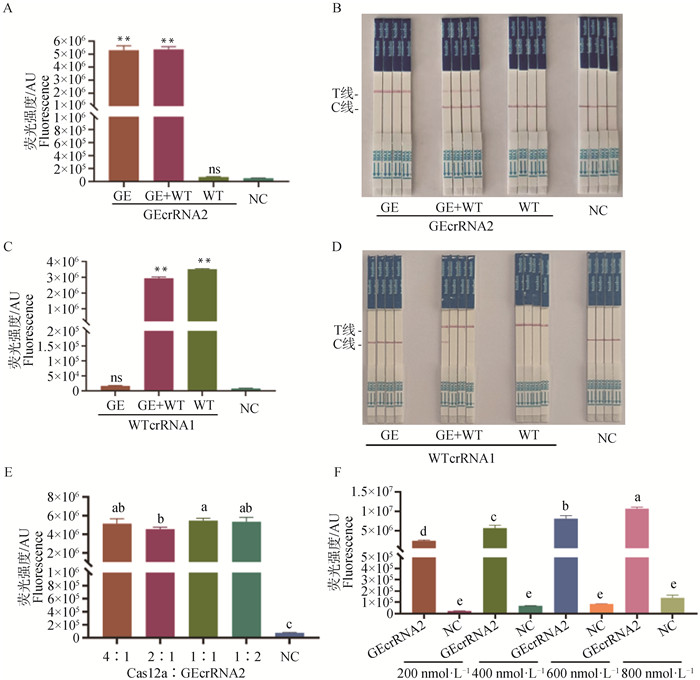
Fig. 4
Sensitivity test of the RPA-CRISPR/Cas12a system A, B. RPA amplification results of gradient-diluted GE and WT plasmid: H2O. Negative control without RPA template; C, D. Sensitivity test of the RPA-CRISPR/Cas12a fluorescence reporter system mediated by GEcrRNA2 and WTcrRNA1; E, F. Sensitivity test of the RPA-CRISPR/Cas12a colloidal gold test strip system mediated by GEcrRNA2 and WTcrRNA1; NC. Negative control without crRNA. Lane 1 to 11 represent 4.1×1010, 4.1×109, 4.1×108, 4.1×107, 4.1×106, 4.1×105, 4.1×104, 4.1×103, 4.1×102, 4.1×101, 4.1×100 copies·μL-1, respectively"
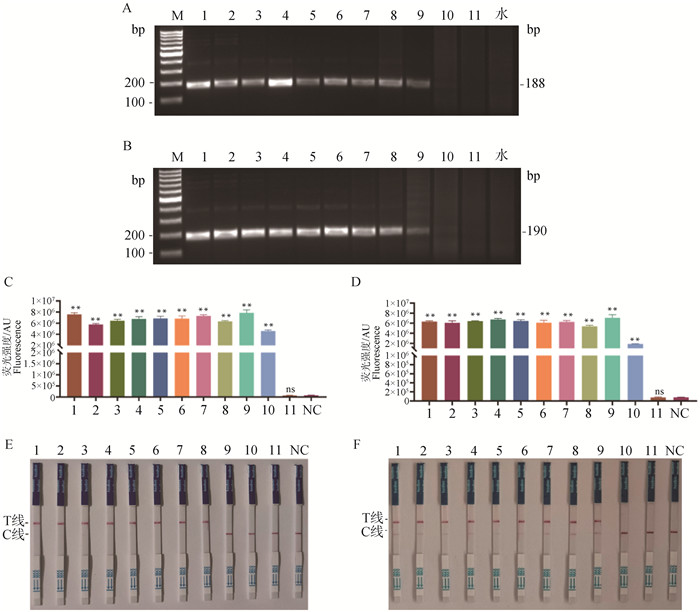
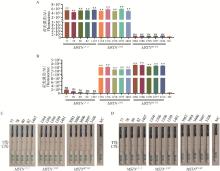
Fig. 5
Validation of the RPA-CRISPR/Cas12a system by using pig ear tissue samples A, B. Detection results of the RPA-CRISPR/Cas12a fluorescence reporter system mediated by GEcrRNA2 and WTcrRNA1; C, D. Detection results of the RPA-CRISPR/Cas12a colloidal gold test strip system mediated by GEcrRNA2 and WTcrRNA1; NC. Negative control without crRNA"
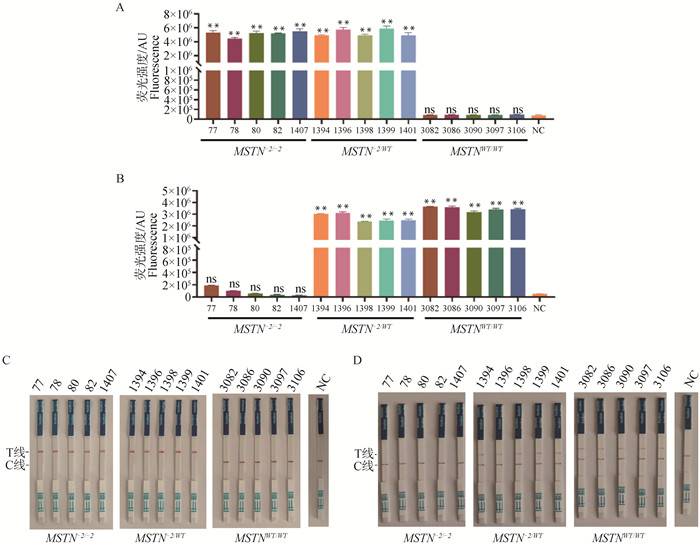
Fig. 6
Application of the RPA-CRISPR/Cas12a system A, B. Detection results of the RPA-CRISPR/Cas12a fluorescence reporter system mediated by GEcrRNA2 and WTcrRNA1; C, D. Detection results of the RPA-CRISPR/Cas12a colloidal gold test strip system mediated by GEcrRNA2 and WTcrRNA1; NC. Negative control without crRNA"

| 1 |
LIU Z G , WU T W , XIANG G M , et al. Enhancing animal disease resistance, production efficiency, and welfare through precise genome editing[J]. Int J Mol Sci, 2022, 23 (13): 7331.
doi: 10.3390/ijms23137331 |
| 2 |
XU K , ZHANG X L , LIU Z G , et al. A transgene-free method for rapid and efficient generation of precisely edited pigs without monoclonal selection[J]. Sci China Life Sci, 2022, 65 (8): 1535- 1546.
doi: 10.1007/s11427-021-2058-2 |
| 3 | 潘东霞, 王辉, 熊本海, 等. CRISPR-Cas9基因编辑技术在牛、羊生产中的应用研究进展[J]. 中国畜牧兽医, 2024, 51 (11): 4880- 4889. |
| PAN D X , WANG H , XIONG B H , et al. Research progress on CRISPR-Cas9 gene editing technology in cattle and sheep production[J]. China Animal Husbandry & Veterinary Medicine, 2024, 51 (11): 4880- 4889. | |
| 4 | 刘志国, 黄雷, 杨丽景, 等. 基因编辑技术在猪生物育种中的研究进展[J]. 中国畜禽种业, 2024, 20 (1): 19- 28. |
| LIU Z G , HUANG L , YANG L J , et al. Research progress of gene editing technology in pig breeding[J]. The Chinese Livestock and Poultry Breeding, 2024, 20 (1): 19- 28. | |
| 5 | 曹慧, 韩博, 孙东晓. 基因编辑技术及其在畜禽遗传育种中的应用研究进展[J]. 中国畜牧杂志, 2024, 60 (7): 6- 12. |
| CAO H , HAN B , SUN D X . Research progress on gene editing technology and its application in livestock and poultry genetic breeding[J]. Chinese Journal of Animal Science, 2024, 60 (7): 6- 12. | |
| 6 |
BURKARD C , LILLICO S G , REID E , et al. Precision engineering for PRRSV resistance in pigs: Macrophages from genome edited pigs lacking CD163 SRCR5 domain are fully resistant to both PRRSV genotypes while maintaining biological function[J]. PLoS Pathog, 2017, 13 (2): e1006206.
doi: 10.1371/journal.ppat.1006206 |
| 7 |
PRATHER R S , WELLS K D , WHITWORTH K M , et al. Knockout of maternal CD163 protects fetuses from infection with porcine reproductive and respiratory syndrome virus (PRRSV)[J]. Sci Rep, 2017, 7 (1): 13371.
doi: 10.1038/s41598-017-13794-2 |
| 8 | WELLS K D , BARDOT R , WHITWORTH K M , et al. Replacement of porcine CD163 scavenger receptor cysteine-rich domain 5 with a CD163-like homolog confers resistance of pigs to genotype 1 but not genotype 2 porcine reproductive and respiratory syndrome virus[J]. J Virol, 2017, 91 (2): e01521- 16. |
| 9 | BURKARD C , OPRIESSNIG T , MILEHAM A J , et al. Pigs lacking the scavenger receptor cysteine-rich domain 5 of CD163 are resistant to porcine reproductive and respiratory syndrome virus 1 infection[J]. J Virol, 2018, 92 (16): e00415- 18. |
| 10 | 魏迎辉, 刘志国, 徐奎, 等. CD163双等位基因编辑猪的制备与传代[J]. 中国农业科学, 2018, 51 (4): 770- 777. |
| WEI Y H , LIU Z G , XU K , et al. Generation and propagation of cluster of differentiation 163 biallelic gene editing pigs[J]. Scientia Agricultura Sinica, 2018, 51 (4): 770- 777. | |
| 11 |
WHITWORTH K M , ROWLAND R R R , PETROVAN V , et al. Resistance to coronavirus infection in amino peptidase N-deficient pigs[J]. Transgenic Res, 2019, 28 (1): 21- 32.
doi: 10.1007/s11248-018-0100-3 |
| 12 |
LUO L , WANG S H , ZHU L , et al. Aminopeptidase N-null neonatal piglets are protected from transmissible gastroenteritis virus but not porcine epidemic diarrhea virus[J]. Sci Rep, 2019, 9 (1): 13186.
doi: 10.1038/s41598-019-49838-y |
| 13 |
XU K , ZHOU Y R , MU Y L , et al. CD163 and pAPN double-knockout pigs are resistant to PRRSV and TGEV and exhibit decreased susceptibility to PDCoV while maintaining normal production performance[J]. Elife, 2020, 9, e57132.
doi: 10.7554/eLife.57132 |
| 14 | WANG K K , TANG X C , LIU Y , et al. Efficient generation of orthologous point mutations in pigs via CRISPR-assisted ssODN-mediated homology-directed repair[J]. Mol Ther Nucleic Acids, 2016, 5 (11) |
| 15 | 曾智杰. MSTN基因编辑猪产肉性状和骨骼肌肌纤维类型研究[D]. 合肥: 安徽农业大学, 2020. |
| ZENG Z J. Characteristics of meat production traits and skeletal muscle fiber types of MSTN gene editing pigs[D]. Hefei: Anhui Agricultural University, 2020. (in Chinese) | |
| 16 |
QIAN L L , TANG M X , YANG J Z , et al. Targeted mutations in myostatin by zinc-finger nucleases result in double-muscled phenotype in Meishan pigs[J]. Sci Rep, 2015, 5 (1): 14435.
doi: 10.1038/srep14435 |
| 17 |
REN H Y , XIAO W , QIN X L , et al. Myostatin regulates fatty acid desaturation and fat deposition through MEF2C/miR222/SCD5 cascade in pigs[J]. Commun Biol, 2020, 3 (1): 612.
doi: 10.1038/s42003-020-01348-8 |
| 18 |
FAN Z Y , LIU Z G , XU K , et al. Long-term, multidomain analyses to identify the breed and allelic effects in MSTN-edited pigs to overcome lameness and sustainably improve nutritional meat production[J]. Sci China Life Sci, 2022, 65 (2): 362- 375.
doi: 10.1007/s11427-020-1927-9 |
| 19 | XIANG G H , REN J L , HAI T , et al. Editing porcine IGF2 regulatory element improved meat production in Chinese Bama pigs[J]. Cell Mol Life Sci, 2018, 75 (4): 4619- 4628. |
| 20 |
LIU X F , LIU H B , WANG M , et al. Disruption of the ZBED6 binding site in intron 3 of IGF2 by CRISPR/Cas9 leads to enhanced muscle development in Liang Guang Small Spotted pigs[J]. Transgenic Res, 2019, 28 (1): 141- 150.
doi: 10.1007/s11248-018-0107-9 |
| 21 |
ZU Y , ZHANG X S , REN J F , et al. Biallelic editing of a lamprey genome using the CRISPR/Cas9 system[J]. Sci Rep, 2016, 6, 23496.
doi: 10.1038/srep23496 |
| 22 |
LÓPEZ-VILLAR I , AYALA R , WESSELINK J , et al. Simplifying the detection of MUTYH mutations by high resolution melting analysis[J]. BMC Cancer, 2010, 10, 408.
doi: 10.1186/1471-2407-10-408 |
| 23 |
GUO J G , LI K , JIN L F , et al. A simple and cost-effective method for screening of CRISPR/Cas9-induced homozygous/biallelic mutants[J]. Plant Methods, 2018, 14, 40.
doi: 10.1186/s13007-018-0305-8 |
| 24 |
GOH S K , MUSAFER A , WITKOWSKI T , et al. Comparison of 3 methodologies for genotyping of small deletion and insertion polymorphisms[J]. Clin Chem, 2016, 62 (7): 1012- 1019.
doi: 10.1373/clinchem.2016.256388 |
| 25 |
LI S Y , CHENG Q X , LIU J K , et al. CRISPR-Cas12a has both cis- and trans-cleavage activities on single-stranded DNA[J]. Cell Res, 2018, 28 (4): 491- 493.
doi: 10.1038/s41422-018-0022-x |
| 26 |
CHEN J S , MA E , HARRINGTON L B , et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity[J]. Science, 2018, 360 (6387): 436- 439.
doi: 10.1126/science.aar6245 |
| 27 | 刘晓婷, 李鹏, 左柯铭, 等. 基于CRISPR-Cas12a核酸检测研究进展[J]. 动物医学进展, 2024, 45 (8): 94- 98. |
| LIU X T , LI P , ZUO K M , et al. Progress on CRISPR Cas12 based nucleic acid detections[J]. Progress in Veterinary Medicine, 2024, 45 (8): 94- 98. | |
| 28 | 党生, 张帅, 翟景波. CRISPR/Cas12a系统: 核酸检测的多功能工具[J]. 生物化学与生物物理进展, 2024, 51 (4): 785- 796. |
| DANG S , ZHANG S , ZHAI J B . The versatile tool: CRISPR/Cas12a system for nucleic acid detection[J]. Progress in Biochemistry and Biophysics, 2024, 51 (4): 785- 796. | |
| 29 | 周旭, 王思文, 王秀荣. CRISPR-Cas12a在病原快速检测中的应用[J]. 中国兽医科学, 2022, 52 (8): 1031- 1037. |
| ZHOU X , WANG S W , WANG X R . Application of CRISPR-Cas12a in rapid detection of pathogens[J]. Chinese Veterinary Science, 2022, 52 (8): 1031- 1037. | |
| 30 | 热则古丽·艾科拜尔, 张亚平, 刘浩然, 等. 基于RAA-CRISPR/Cas12a建立牛病毒性腹泻病毒的可视化快速检测方法及其初步应用[J]. 中国兽医科学, 2025, 55 (3): 321- 329. |
| REZEGULI·AIKEBAIER , ZHANG Y P , LIU H R , et al. Establishment and preliminary application of visualization detection technology for bovine viral diarrhea virus based on RAA-CRISPR/Cas12a[J]. Chinese Veterinary Science, 2025, 55 (3): 321- 329. | |
| 31 | 郭心龙, 周勇志, 曹杰, 等. 绵羊无浆体LAMP-CRISPR/Cas12a检测方法的建立[J]. 中国兽医科学, 2025, 55 (3): 330- 337. |
| GUO X L , ZHOU Y Z , CAO J , et al. Development of LAMP-CRISPR/Cas12a assay for the detection of Anaplasmaovis[J]. Chinese Veterinary Science, 2025, 55 (3): 330- 337. | |
| 32 | XIONG D , DAI W J , GONG J J , et al. Rapid detection of SARS-CoV-2 with CRISPR-Cas12a[J]. PLoS Biol, 2020, 18 (12): e3000978. |
| 33 | QU H , ZHANG W J , LI J H , et al. A rapid and sensitive CRISPR-cas12a for the detection of fusobacterium nucleatum[J]. Microbiol Spectr, 2024, 12 (2): e0362923. |
| 34 | MAYURAMART O , NIMSAMER P , RATTANABURI S , et al. Detection of severe acute respiratory syndrome coronavirus 2 and influenza viruses based on CRISPR-Cas12a[J]. Exp Biol Med (Maywood), 2021, 246 (4): 400- 405. |
| 35 | QIN C , LIU J J , ZHU W Q , et al. One-pot visual detection of African swine fever virus using CRISPR-Cas12a[J]. Front Vet Sci, 2022, 9, 962438. |
| 36 | ZHANG K L , SUN Z Y , SHI K D , et al. RPA-CRISPR/Cas12a-based detection of Haemophilus parasuis[J]. Animals (Basel), 2023, 13 (21): 3317. |
| 37 | CAO X Y , CHANG Y B , TAO C Q , et al. Cas12a/Guide RNA-based platforms for rapidly and accurately identifying Staphylococcus aureus and Methicillin-Resistant S. aureus[J]. Microbiol Spectr, 2023, 11 (2): e0487022. |
| 38 | WANG M Y , LIU X J , YANG J T , et al. CRISPR/Cas12a-based biosensing platform for the on-site detection of single-base mutants in gene-edited rice[J]. Front Plant Sci, 2022, 13, 944295. |
| 39 | HUA Y F , WANG C , HUANG J , et al. A simple and efficient method for CRISPR/Cas9-induced mutant screening[J]. J Genet Genomics, 2017, 44 (4): 207- 213. |
| 40 | WANG Z , HUANG C M , WEI S , et al. A CRISPR/Cas12a-mediated sensitive DNA detection system for gene-edited rice[J]. J AOAC Int, 2023, 106 (3): 558- 567. |
| 41 | JANUDIN A A S , KURUP C P , CHEE L Y , et al. Amplification-based CRISPR/Cas12a biosensor targeting the COX1 gene for specific detection of porcine DNA[J]. ACS Omega, 2023, 8 (41): 38212- 38219. |
| 42 | ZHANG H X , ZHANG C X , LU S H , et al. Cas12a-based one-pot SNP detection with high accuracy[J]. Cell Insight, 2023, 2 (2): 100080. |
| 43 | YE X J , LEI B . The current status and trends of DNA extraction[J]. Bioessays, 2023, 45 (8): e2200242. |
| 44 | LIU Y Q , LIN L Y , WEI H G , et al. Design and development of a rapid meat detection system based on RPA-CRISPR/Cas12a-LFD[J]. Curr Res Food Sci, 2023, 7, 100609. |
| 45 | BHATTACHARJEE G , GOHIL N , LAM N L , et al. CRISPR-based diagnostics for detection of pathogens[J]. Prog Mol Biol Transl Sci, 2021, 181, 45- 57. |
| 46 | TANG G Y , ZHANG Z L , TAN W , et al. RT-RPA-Cas12a-based assay facilitates the discrimination of SARS-CoV-2 variants of concern[J]. Sens Actuators B Chem, 2023, 381, 133433. |
| [1] | Lu FENG, Hong TIAN, Haixue ZHENG, Zhengwang SHI, Juncong LUO, Xiaoyang ZHANG, Juanjuan WEI, Jing ZHOU, Huancheng LIAO, Wanying WANG. A Detection Method of African Swine Fever Virus based on Enzymatic Recombinase Amplification [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(9): 4226-4231. |
| [2] | ZHANG Senhao, WANG Xueying, CAI Limeng, XIE Weichun, KUANG Hongdi, WANG Xiaona, LI Jiaxuan, CUI Wen, JIANG Yanping, ZHOU Han, SHAN Zhifu, WANG Li, QIAO Xinyuan, LI Yijing, TANG Lijie. Establishment and Application of Rapid Diagnosis Method for RPA of Porcine Epidemic Diarrhea Virus [J]. Acta Veterinaria et Zootechnica Sinica, 2023, 54(6): 2498-2508. |
| [3] | GAO Pingping, FU Jinyu, WANG Liyang, SHI Shuobo, ZHANG Yueping, ZHANG Di. Rapid RPA-CRISPR/Cas12a Detection Platform for Dermatophytes in Dogs and Cats [J]. Acta Veterinaria et Zootechnica Sinica, 2023, 54(11): 4702-4711. |
| [4] | HUANG Jian, LIU Yunjia, YANG Xiaonong, LI Yan. RPA-Cas12a-LFD Based Nucleic Acid Detection for Feline Herpesvirus-1 and Preliminary Application [J]. Acta Veterinaria et Zootechnica Sinica, 2022, 53(5): 1638-1643. |
| [5] | ZHANG Shanshan, HE Bin, LI Shuguang, LIU Mingcheng, JIANG Jinqing, HU Jianhe, LEI Liancheng, SHEN Zhiqiang, XIA Xiaojing. Rapid Detection of Streptococcus suis with Visual RPA-LFD Technology [J]. Acta Veterinaria et Zootechnica Sinica, 2022, 53(2): 538-547. |
| [6] | XU Lu, ZHANG Qianyi, XIA Yingju, WANG Zhen, LI Cui, ZOU Xingqi, WANG Qin, ZHAO Qizu. Result Analysis of National Proficiency Testing Program for the Detection of Classical Swine Fever Virus Nucleic Acid [J]. Acta Veterinaria et Zootechnica Sinica, 2020, 51(8): 1949-1955. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||