





畜牧兽医学报 ›› 2025, Vol. 56 ›› Issue (9): 4718-4729.doi: 10.11843/j.issn.0366-6964.2025.09.048
孙品之( ), 屈盈盈, 张沁, 杨丽雯, 李彦歌, 张怡清清, 张雨, 路浩*(
), 屈盈盈, 张沁, 杨丽雯, 李彦歌, 张怡清清, 张雨, 路浩*( )
)
收稿日期:2024-11-11
出版日期:2025-09-23
发布日期:2025-09-30
通讯作者:
路浩
E-mail:sunpinzhi@nwafu.edu.cn;luhao@nwsuaf.edu.cn
作者简介:孙品之(1998-),女,陕西西安人,硕士生,主要从事植物毒素生物合成与调控研究,E-mail: sunpinzhi@nwafu.edu.cn
基金资助:
SUN Pinzhi( ), QU Yingying, ZHANG Qin, YANG Liwen, LI Yange, ZHANG Yiqingqing, ZHANG Yu, LU Hao*(
), QU Yingying, ZHANG Qin, YANG Liwen, LI Yange, ZHANG Yiqingqing, ZHANG Yu, LU Hao*( )
)
Received:2024-11-11
Online:2025-09-23
Published:2025-09-30
Contact:
LU Hao
E-mail:sunpinzhi@nwafu.edu.cn;luhao@nwsuaf.edu.cn
摘要:
本研究旨在利用CRISPR/Cas9基因编辑技术研究p5cr(吡咯啉-5-羧酸还原酶)在金龟子绿僵菌合成苦马豆素中的作用,为苦马豆素的生物合成和工业化生产提供理论依据。试验以金龟子绿僵菌(Metarhizium anisopliae)为研究对象,使用qRT-PCR方法检测苦马豆素合成通路基因p5cr、sdh、pks和p450的转录水平,并对发酵培养1~7 d的金龟子绿僵菌的苦马豆素产量和相关基因表达量进行相关性分析,筛选出p5cr基因作为目标基因,采用CRISPR/Cas9基因编辑技术对催化酶基因p5cr进行敲除和过表达,验证p5cr基因与金龟子绿僵菌合成苦马豆素的相关性。结果显示,苦马豆素的产量与pks基因表达量之间为强相关性,而与p5cr、sdh和p450基因表达量之间均为极强相关性;敲除和过表达金龟子绿僵菌p5cr基因会导致苦马豆素产量显著降低(P<0.01)和升高(P<0.001)。上述研究结果表明,p5cr基因与金龟子绿僵菌的苦马豆素合成具有极强相关性,并且该基因在金龟子绿僵菌合成苦马豆素通路中起重要调节作用,为后续深入研究p5cr基因的功能及其调控作用奠定基础。
中图分类号:
孙品之, 屈盈盈, 张沁, 杨丽雯, 李彦歌, 张怡清清, 张雨, 路浩. p5cr基因在金龟子绿僵菌合成苦马豆素中相关性分析[J]. 畜牧兽医学报, 2025, 56(9): 4718-4729.
SUN Pinzhi, QU Yingying, ZHANG Qin, YANG Liwen, LI Yange, ZHANG Yiqingqing, ZHANG Yu, LU Hao. Correlation Analysis of the p5cr Gene in Swainsonine Biosynthesis in Metarhizium anisopliae[J]. Acta Veterinaria et Zootechnica Sinica, 2025, 56(9): 4718-4729.
表 1
qRT-PCR引物"
| 基因 Gene | 序列(5′→3′) Sequence |
| sdh-F | GCCCAACGCTACAACATC |
| sdh-R | CTCCAAGAAGCCGACCT |
| p5cr-F | GCGTATTGGTGATGTTGTC |
| p5cr-R | CTCGCATGGATTGAGTAG |
| pks-F | TACACTGGCGTCTGGATA |
| pks-R | CGGGAAGTAGTTTGGGTC |
| p450-F | ACATTGCCCTCCTTTACC |
| p450-R | TGCGCCAGTCACGAACA |
| ITS-F | AGCGAAATGCGATAAGTAATG |
| ITS-F | GACGGCTGTGCTGGAAAA |
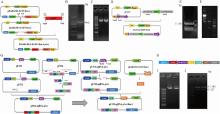
图 2
构建载体 A. 过表达载体pBARGPE1-p5cr的构建;B. 通过PCR鉴定带有目标基因的pBARGPE1-p5cr载体;C. 通过PCR鉴定过表达载体的单克隆菌落;D. CRISPR-HDR介导的p5cr敲除载体的供体构建;E. 通过PCR验证up-Kozak-EGEP-down片段;F. 供体pUC19-up-EGEP-down验证;G. CRISPR/Cas9 p5cr敲除载体pF3;H. pF332-sgRNA-p5cr-BenA的关键因子;I. 中间载体质粒pF332-sgRNA-p5cr(16 513 bp)线性化片段的验证;J. 对pF332-sgRNA-p5cr-BenA载体进行酶切并通过PCR验证"
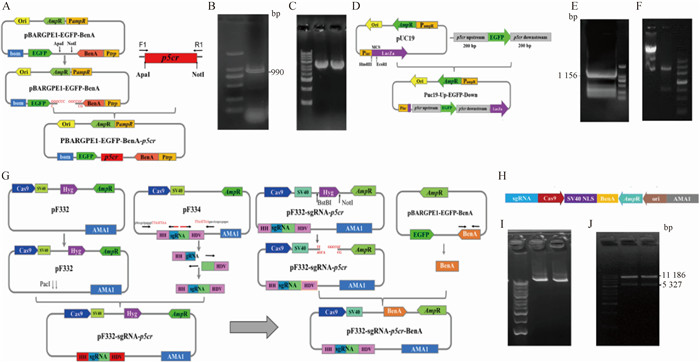
表 2
用于载体构建和验证的引物"
| 引物 Primer | 序列 Sequence (5′→3′) | 用途 Purpose |
| p5cr-F | TCCGCTTTCGCCTTTCTCA | 扩增p5cr Amplifying p5cr |
| p5cr-R | CTGAAATTGCCCATCTCAACg | |
| p5cr-ApaⅠ | tcgacctcgagggg$\underline{{\rm{GGGCCC}}}$GGTCCGCTTTCGCCTTTCTC | 构建pBARGPE1-p5cr Construct pBARGPE1-p5cr |
| p5cr1 | ataaataacgtcatCTGAAATTGCCCATCTCAACGGGC | |
| p5cr2 | tgggcaatttcagATGACGTTATTTATGAGATGGG | |
| p5cr-NotⅠ | aagggttccggatc$\underline{{\rm{GCGGCCGC}}}$TCGACC | |
| EGFP-F3 | AGAAGCGCGATCACATGGTC | pBARGPE1-p5cr阳性转化子鉴定引物 Primers for identification of pBARGPE1-p5cr positive transformants |
| p5cr-R3 | TGCAGAGGCTCAGAGAATGG | |
| EGFP-F | ATGGTGAGCAAGGGCGAGGAGCTGTT | 扩增EGFP Amplifying EGFP |
| EGFP-R | TTACTTGTACAGCTCGTCCATGCCGAGAGTGATC | |
| sgRNA-PacI-F | gtttccgctgagggt$\underline{{\rm{TTAATTAA}}}$TGCGTAAGCTCCCTAATTGGCCC | 构建pF332-sgRNA-p5cr-BenA Construct pF332-sgRNA-p5cr-BenA |
| SgRNA-PacI-R | GTCTCGGCTGAGGTC$\underline{{\rm{TTAATT}}}$aatgagccaagagcggattcctcag | |
| F2 | aagcgcaaggtctgaATGCGTGAGATTGTCCATCTTC | |
| R2 | gtcggcatctactCTATTCCTCGCCCTCAATTGGGACCTC | |
| F3 | atagagtagatgccgacCGATCCCCACTACCGC | |
| R3 | attaaaccctcagcGCGGCCGCATTTA | |
| F4 | tggagcgctccagcTTCGAAAAG | |
| R4 | acaatctcacgcatTCAGACCTTGCGCTTCTTC | |
| Cas-F | TCCGACAATGTTCCAAGCGA | pF332-sgRNA-p5cr载体鉴定引物Primers for identification of pF332-sgRNA-p5cr vectors |
| Cas-R | TCGCCGTTGGTCTCAATCAA | |
| pFC332-F | GAAGATCTCATGGTCATAGCTG | pF332-sgRNA-p5cr-BenA载体鉴定引物 Primers for identification of pF332-sgRNA-p5cr-BenA vectors |
| p5cr-sgRNA-R | GATGGAGTATGGATGTAGCAAAGT | |
| Cas9-F1 | TGATGCGACCCTGATTCATC | |
| Ben-R1 | ACGGAAGAGTTGACCGAAAG | |
| Up-F1 | gacggccagtCGATCGATGGAGACTTACTTGGCGTTA | 构建up-kozak-EGEP-down Construct up-kozak-EGEP-down |
| Down-R1 | atgattacgccGTCTATCTAACCTAAAACCACCGC | |
| up-kozak-EGFP-F | aaccagtcgtgttgaacctggttc$\underline{{\rm{GCCACC}}}$ATGGTGAGCAAGGGCGAGGAGCTGTT | |
| EGFP-Down-R | gatcactctcggcatggacgagctgtacaagtAATGAG AATGGCAATTCCCATGGTGCC | |
| Cas-F | TCCGACAATGTTCCAAGCGA | Δp5cr载体验证引物 Primers for Δp5cr vector verification |
| Cas-R | TCGCCGTTGGTCTCAATCAA | |
| UP-F2 | ATGGAGACTTACTTGGCGTTA | |
| p5cr-R | ATGTGCAATACTGCTGCTCTT | |
| EGFP-R | TTACTTGTACAGCTCGTCCATGCCGAGAGTGATC |
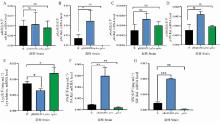
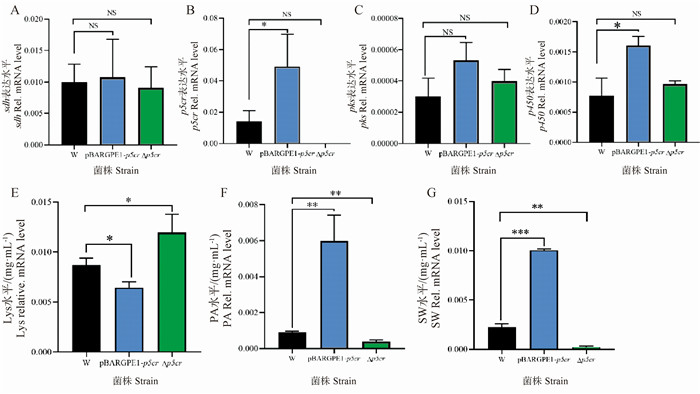
图 9
W、PBARGPE1-p5cr和Δp5cr菌株的基因表达检测和苦马豆素、哌可酸、赖氨酸含量的测定 A. W、pBARGPE1-p5cr和Δp5cr菌株中sdh基因的表达水平;B. W、pBARGPE1-p5cr和Δp5cr菌株中p5cr基因的表达水平;C. W、pBARGPE1-p5cr和Δp5cr菌株中pks基因的表达水平;D. W、pBARGPE1-p5cr和Δp5cr菌株中p450基因的表达水平;E. W、pBARGPE1-p5cr和Δp5cr菌株中的Lys水平;F. W、pBARGPE1-p5cr和Δp5cr菌株中的PA水平;G. W、pBARGPE1-p5cr和Δp5cr菌株中的苦马豆素水平"
| 1 |
FUK Y,CHENX,SHOUN,et al.Swainsonine induces liver inflammation in mice via disturbance of gut microbiota and bile acid metabolism[J].J Agric Food Chem,2023,71,1758-1767.
doi: 10.1021/acs.jafc.2c08519 |
| 2 |
WANGS,GUOY Z,YANGC,et al.Swainsonine triggers paraptosis via ER stress and mapk signaling pathway in rat primary renal tubular epithelial cells[J].Front Pharmacol,2021,12,715285.
doi: 10.3389/fphar.2021.715285 |
| 3 |
LIUY L,ZHANGS H,WANGW N,et al.Swainsonine-induced vacuolar degeneration is regulated by mTOR-mediated autophagy in HT22 cells[J].Toxicol Lett,2023,373,41-52.
doi: 10.1016/j.toxlet.2022.11.007 |
| 4 |
SILVEIRAC R F,CIPELLIM,MANZINEC,et al.Swainsonine, an alpha-mannosidase inhibitor, may worsen cervical cancer progression through the increase in myeloid derived suppressor cells population[J].PLoS One,2019,14,e0213184.
doi: 10.1371/journal.pone.0213184 |
| 5 |
RENS E,FANC,ZHANGL Z,et al.The plant secondary compound swainsonine reshapes gut microbiota in plateau pikas (Ochotona curzoniae)[J].Appl Microbiol Biotechnol,2021,105,6419-6433.
doi: 10.1007/s00253-021-11478-6 |
| 6 |
LUH,QUANH Y,RENZ H,et al.The genome of Undifilum oxytropis provides insights into swainsonine biosynthesis and locoism[J].Sci Rep,2016,6,30760.
doi: 10.1038/srep30760 |
| 7 |
COOKD,DONZELLIB G G,CREAMERR,et al.Swainsonine biosynthesis genes in diverse symbiotic and pathogenic fungi[J].G3 (Bethesda),2017,7,1791-1797.
doi: 10.1534/g3.117.041384 |
| 8 |
LID,ZHAOX L,LUP,et al.The Effects of swnH1 gene function of endophytic fungus Alternaria oxytropis OW 7.8 on its swainsonine biosynthesis[J].Microorganisms,2024,12(10):2081.
doi: 10.3390/microorganisms12102081 |
| 9 |
LUOF F,HONGS,CHENB,et al.Unveiling of swainsonine biosynthesis via a multibranched pathway in fungi[J].ACS Chem Biol,2020,15(9):2476-2484.
doi: 10.1021/acschembio.0c00466 |
| 10 | 孙璐. PKS基因在金龟子绿僵菌合成苦马豆素中的作用研究[D]. 杨凌: 西北农林科技大学, 2021. |
| SUN L. The effect on PKS gene in swainsonine synthesis of Metarhizium anisopliae[D]. Yangling: Northwestern Agriculture and Forestry University, 2021. (in Chinese) | |
| 11 |
BROQUISTH,MASONP,HAGLERW,et al.Identification of swainsonine as a probable contributory mycotoxin in moldy forage mycotoxicosis[J].Appl Environ Microbiol,1984,48,386-388.
doi: 10.1128/aem.48.2.386-388.1984 |
| 12 |
张雨,朱燕丽,李博,等.金龟子绿僵菌发酵液中苦马豆素含量及催化酶基因mRNA表达分析[J].畜牧兽医学报,2020,51(4):881-887.
doi: 10.11843/j.issn.0366-6964.2020.04.025 |
|
ZHANGY,ZHUY L,LIB,et al.mRNA expression analysis of catalytic enzyme gene and content of swainsonine in the fermentation broth of Metarhizium anisopliae[J].Acta Veterinaria et Zootechnica Sinica,2020,51(4):881-887.
doi: 10.11843/j.issn.0366-6964.2020.04.025 |
|
| 13 |
孙璐,宋润杰,路浩,等.SWNR基因在金龟子绿僵菌合成苦马豆素中的作用[J].畜牧兽医学报,2021,52(5):1439-1446.
doi: 10.11843/j.issn.0366-6964.2021.05.030 |
|
SUNL,SONGR J,LUH,et al.The role of SWNR gene on the biosynthetic pathway of the swainsonine in Metarhizium anisopliae[J].Acta Veterinaria et Zootechnica Sinica,2021,52(5):1439-1446.
doi: 10.11843/j.issn.0366-6964.2021.05.030 |
|
| 14 | GUOC C,ZHANGL,ZHAOQ Q,et al.Host-species variation and environment influence endophyte symbiosis and mycotoxin levels in Chinese Oxytropis species[J].Toxins,2022,14,18. |
| 15 |
SCHMITTGENT D,LIVAKK J.Analyzing real-time PCR data by the comparative C(T) method[J].Nat Protoc,2008,3,1101-1108.
doi: 10.1038/nprot.2008.73 |
| 16 |
NODVIGC S,NIELSENJ B,KOGLEM E,et al.A CRISPR-Cas9 system for genetic engineering of filamentous fungi[J].PLoS One,2015,10,e0133085.
doi: 10.1371/journal.pone.0133085 |
| 17 |
HUANGE X,ZHANGY,SUNL,et al.swnk plays an important role in the biosynthesis of swainsonine in Metarhizium anisopliae[J].Biotechnol Lett,2023,45,509-519.
doi: 10.1007/s10529-023-03356-0 |
| 18 | CHENY Y,CAIC L,YANGJ F,et al.Development of the CRISPR/Cas9 system for the marine-derived fungi Spiromastix sp. SCSIO F190 and Aspergillus sp. SCSIO SX7S7[J].J Fungi(Basel),2022,8(7):715. |
| 19 |
DINGQ,YEC.Microbial cell factories based on filamentous bacteria, yeasts, and fungi[J].Microb Cell Fact,2023,22(1):20.
doi: 10.1186/s12934-023-02025-1 |
| 20 |
DICARLOJ E,NORVILLEJ E,MALIP,et al.Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems[J].Nucleic Acids Res,2013,41,4336-4343.
doi: 10.1093/nar/gkt135 |
| 21 |
POHLC,KIELJ A K W,DRIESSENA J M,et al.CRISPR/Cas9 based genome editing of Penicillium chrysogenum[J].ACS synth biol,2016,5,754-764.
doi: 10.1021/acssynbio.6b00082 |
| 22 |
CLEMMENSENS E,KROMPHARDTK J K,FRANDSENR J N.Marker-free CRISPR-Cas9 based genetic engineering of the phytopathogenic fungus, Penicillium expansum[J].Fungal Genet Biol,2022,160,103689.
doi: 10.1016/j.fgb.2022.103689 |
| 23 |
SHIT Q,GAOJ,WANGW J,et al.CRISPR/Cas9-based genome editing in the filamentous fungus Fusarium fujikuroi and its application in strain engineering for gibberellic acid production[J].ACS Synth Biol,2019,8,445-454.
doi: 10.1021/acssynbio.8b00478 |
| 24 | LIUR,CHENL,JIANGY P,et al.Efficient genome editing in filamentous fungus Trichoderma reesei using the CRISPR/Cas9 system[J].Cell Discov,2015,1,15007. |
| 25 |
SCHIMMELJ,MUÑOZ-SUBIRANAN,KOOLH,et al.Modulating mutational outcomes and improving precise gene editing at CRISPR-Cas9-induced breaks by chemical inhibition of end-joining pathways[J].Cell Rep,2023,42,112019.
doi: 10.1016/j.celrep.2023.112019 |
| 26 |
AMANOM,MIZUGUCHIH,SANOT,et al.Recombinant expression, molecular characterization and crystal structure of antitumor enzyme, L-lysine α-oxidase from Trichoderma viride[J].J Biochem,2015,157(6):549-59.
doi: 10.1093/jb/mvv012 |
| 27 |
JIANGC M,LVG B,TUY Y,et al.Applications of CRISPR/Cas9 in the synthesis of secondary metabolites in filamentous fungi[J].Front Microbiol,2021,12,638096.
doi: 10.3389/fmicb.2021.638096 |
| 28 |
LIUW W,ANC Y,SHUX,et al.A dual-plasmid CRISPR/Cas system for mycotoxin elimination in polykaryotic industrial fungi[J].ACS Synth Biol,2020,9(8):2087-2095.
doi: 10.1021/acssynbio.0c00178 |
| 29 |
ZHANGL,WUR L,MURL A J,et al.Assembly of high-quality genomes of the locoweed Oxytropis ochrocephala and its endophyte Alternaria oxytropis provides new evidence for their symbiotic relationship and swainsonine biosynthesis[J].Mol Ecol Resour,2023,23,253-272.
doi: 10.1111/1755-0998.13695 |
| 30 |
LIANGY,LIS W,SONGX D,et al.Swainsonine producing performance of Alternaria oxytropis was improved by heavy-ion mutagenesis technology[J].FEMS Microbiol Lett,2021,368,fnab047.
doi: 10.1093/femsle/fnab047 |
| [1] | 余永涛, 毛彦妮, 赵清梅, 李金荣, 白晓南, 薛佳祺, 张浩东. 甲基磺酸乙酯诱变的棘豆链格孢菌菌株苦马豆素合成基因簇相关基因表达模式分析[J]. 畜牧兽医学报, 2022, 53(4): 1241-1251. |
| [2] | 王维夫, 钱亚光, 卢萍, 何珊, 杜玲, 李玉玲, 高峰. 内蒙古不同小花棘豆种群苦马豆素及其内生真菌关系的分析[J]. 畜牧兽医学报, 2022, 53(1): 304-314. |
| [3] | 孙璐, 宋润杰, 路浩, 王敬龙, 莫重辉, 赵宝玉. swnR基因在金龟子绿僵菌合成苦马豆素中的作用[J]. 畜牧兽医学报, 2021, 52(5): 1439-1446. |
| [4] | 张雨, 朱燕丽, 李博, 孙璐, 宋润杰, 黄恩霞, 赵宝玉, 路浩. 金龟子绿僵菌发酵液中苦马豆素含量及催化酶基因mRNA表达分析[J]. 畜牧兽医学报, 2020, 51(4): 881-887. |
| [5] | 李中央, 王圣翔, 强煜云, 田甜, 赵宝玉, 吴晨晨. 苦马豆素抑制N-聚糖加工对生殖激素分泌功能的影响[J]. 畜牧兽医学报, 2020, 51(12): 3171-3180. |
| [6] | 余永涛, 李金荣, 赵清梅, 何生虎, 陈娟, 杨奇, 毛彦妮. 直立黄芪中产苦马豆素真菌的分离与鉴定[J]. 畜牧兽医学报, 2018, 49(8): 1770-1780. |
| [7] | 黄鑫,梁剑平,高旭东,郝宝成. 苦马豆素的来源、药理作用及检测方法研究进展[J]. 畜牧兽医学报, 2016, 47(6): 1075-1085. |
| [8] | 张蕾蕾,余永涛,何生虎,赵清梅,葛松. 不同因素对疯草内生真菌合成苦马豆素的影响[J]. 畜牧兽医学报, 2015, 46(1): 163-173. |
| [9] | 杨晓雯,赵宝玉,路浩,白玛桑姆,曹丹丹,周启武. 不同培养条件对甘肃棘豆内生真菌多样性的影响[J]. 畜牧兽医学报, 2013, 44(10): 1660-1666. |
| [10] | 陈基萍, 赵宝玉, 路浩, 丁来弟, 张广芳, 李小蒙, 高源, 周启武, 杨晓雯. 中国主要疯草内生真菌分离鉴定及其抑菌活性研究[J]. 畜牧兽医学报, 2012, 43(9): 1471-1478. |
| [11] | 卢围;路浩;赵宝玉;荣杰;陈基萍;王瑞;高文超;陈占莉. 小花棘豆产苦马豆素内生真菌的筛选与鉴定[J]. 畜牧兽医学报, 2011, 42(3): 429-436. |
| [12] | 马尧;路浩;赵宝玉;来航线;李蓉;王占新;陈基萍;卢围;荣杰. 产苦马豆素真菌的筛选与鉴定[J]. 畜牧兽医学报, 2010, 41(5): 621-629. |
| [13] | 李勤凡;王建华;李 蓉;陈 亚;刘志滨. 冰川棘豆生物碱分析及苦马豆素的分离、鉴定[J]. 畜牧兽医学报, 2005, 36(12): 1339-1343. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
